Study results confirm significant heritability for feed efficiency traits in a Nile tilapia breeding population

One of the key features of aquaculture species compared with terrestrial farmed species is their greater feed efficiency (FE). For example, fish need around six times less feed than cattle to produce the same amount of body mass. However, feed remains the primary cost for farmed fish production and relatively little direct selection for improved feed efficiency has been performed for most aquaculture species. Therefore, genetic improvement of FE would enhance the economic sustainability of aquaculture and reduce environmental impacts, including greenhouse gas emissions.
Since the domestication of aquaculture species is relatively young, selective breeding offers a great potential to improve commercially important traits. The benefits of genetic improvement have been illustrated in some key aquaculture species in relation to growth rate and disease resistance, particularly when augmented by genomic tools. However, comparatively little direct focus has been placed on feed efficiency in most aquaculture species, most likely due to the challenges of measuring feed intake efficiently and accurately at an individual level.
The Nile tilapia (Oreochromis niloticus) is the third-most produced farmed species globally. Farmed across a wide range of production systems, this species is considered a critical protein source for human consumption in undeveloped and developing countries. Recently, a wide variety of genomic tools have been developed for Nile tilapia and recently reviewed in Yáñez et al.
These genomic tools have facilitated studies to unravel the relationships among improved strains, to detect regions associated with important traits, and to assess the accuracy of prediction of breeding values using genomic selection (GS) models, compared with pedigree-based models. However, there is a lack of studies targeting the identification of genomic regions associated with FE traits (i.e., those which involve the recording of feed intake), or assessing the potential impact of using genomic approaches to improve these traits in Nile tilapia breeding populations.
This article – adapted and summarized from the original publication [Barría, A. et al. 2021. Genomic Selection and Genome-wide Association Study for Feed-Efficiency Traits in a Farmed Nile Tilapia (Oreochromis niloticus) Population. Front. Genet. 12:737906 – evaluated the genetic architecture in a Nile tilapia breeding population to assess genomic prediction for feed-efficiency traits in this species.
Study setup
This study used tissue samples we archived from a previous study of genetic parameters of FE-related traits in GIFT tilapia. The fish used were from the Genetically Improved Farmed Tilapia (GIFT) breeding program based in Jitra, Malaysia, and managed by WorldFish. These fish had been selected for increased growth rate for 15 generations in Malaysia at the time the experiment was performed.
Data used included recorded feed conversion ratio (FCR), body weight gain (BWG), residual feed intake (RFI) and feed intake (FI) traits for 40 full-sibling [same biological parents] families from the GIFT Nile tilapia breeding population. For detailed information on the feed conversion experimental challenge; trait definitions; various genetic analyses and predictions of breeding values; refer to the original publication.
Evaluating genetic parameters for resistance to Tilapia Lake Virus in Nile tilapia
Results and discussion
Our study generated genotype data from tissue samples collected from a previous study we carried out in which genetic parameters for FE traits were estimated in a Nile tilapia breeding population. Through quantitative genetic analyses and using pedigree data with information from up to 15 generations, significant genetic variation was detected for all the assessed traits, FCR, BWG, RFI and FI.
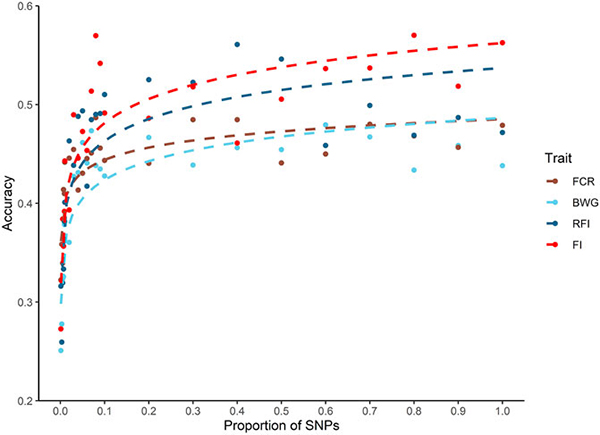
Regarding heritability and genetic correlations, we estimated significant heritabilities, ranging from 0.12 to 0.22, for all the assessed traits. Genetic-parameters estimates for FE traits are relatively rare in aquaculture species, but have been extensively assessed in livestock species with significant heritabilities detected, ranging from 0.13 up to 0.84. Some studies on aquaculture species have also detected moderate to high heritabilities for average daily gain (ADG), daily feed intake (DFI), feed efficiency ratio (FER) and RFI in a breeding Pacific white shrimp (Litopenaeus vannamei) population.
A moderate genetic correlation was found between BWG and both FCR and RFI. This favorable and significant correlation suggests that selection for BWG (a simple trait to measure) would result in favorable correlated responses for feed efficiency traits within the current population. Although these results agree with previous studies in sea bass and rainbow trout, they contrast with other studies which found no significant correlations. It is challenging to explain this discrepancy between the pedigree and the genomic data.
With few exceptions, commercially important traits in aquaculture are underpinned by a polygenic genetic architecture [affected by many different genes]. This genetic architecture is also typical of FE traits in several breeding populations of livestock. Various authors have reported a polygenic architecture for these traits for other species like Atlantic salmon. Our results agree with these previous studies, suggesting a polygenic architecture for feed efficiency traits in this current GIFT Nile tilapia breeding population.
Regarding genomic prediction and impact of SNP density – since several factors such as heritability, population size, and genetic architecture underlying traits can impact the accuracy of the genomic predictions – it is necessary to compare the performance of different models when a trait is analyzed for the first time within a specific population. To our knowledge, ours here is the first study to assess genomic predictions for feed efficiency traits in Nile tilapia.
The impact of SNP density on predicted accuracies has been studied thoroughly for a wide variety of traits and species in aquaculture. The results of these studies highlight the potential to significantly reduce SNP density without affecting the accuracy of prediction. Our study shows that the accuracy of breeding value prediction using the genomic data was up to 34 percent higher than using pedigree records. An SNP density of approximately 5,000 SNPs was sufficient to achieve similar prediction accuracy as the full genotype data set. Thus, breeding programs could potentially use low-density SNP markers (typically cheaper than high-density panels) to achieve higher accuracies than using pedigree-based models, potentially increasing genetic gain in a cost-efficient manner.
Perspectives
Our results confirmed significant heritabilities for feed-efficiency traits in a Nile tilapia breeding population, highlighting that genetic improvement is feasible. A negative, but favorable, genetic correlation was detected between BWG and FCR using the genomic data, suggesting selection for BWG may indirectly improve FCR. The traits were polygenic, suggesting the genomic selection may be the most effective route to incorporating genotype data into selection decisions.
Genomic prediction markedly outperformed pedigree-based prediction, and this was the case even for relatively low-density SNP markers. Overall, our study highlights the potential for genomic selection to improve feed efficiency traits in Nile tilapia breeding programs.
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Authors
-
Agustin Barría, Ph.D.
The Roslin Institute and Royal (Dick) School of Veterinary Studies
University of Edinburgh Easter Bush
Midlothian, United Kingdom -
John A.H. Benzie, Ph.D.
WorldFish, Bayan Lepas, Malaysia; and
School of Biological Earth and Environmental Sciences
University College Cork
Cork, Ireland -
Prof. Ross D. Houston, Ph.D.
The Roslin Institute and Royal (Dick) School of Veterinary Studies
University of Edinburgh Easter Bush
Midlothian, United Kingdom -
Prof. Dirk-Jan De Koning, Ph.D.
Department of Animal Breeding and Genetics
Swedish University of Agricultural Sciences
Uppsala, Sweden -
Hugues de Verdal, Ph.D.
Corresponding author
CIRAD, UMR ISEM
Montpellier, France; and
UMR AGAP Institut
Univ. Montpellier
CIRAD, INRAE, Institut Agro
Montpellier, France[114,102,46,100,97,114,105,99,64,108,97,100,114,101,118,95,101,100,46,115,101,117,103,117,104]
Tagged With
Related Posts

Responsibility
‘Model’ tilapia venture shows mettle in Mozambique
On the shores of Lake Cahora Bassa, Chicoa Fish Farm hopes to create a ripple effect to improve fish supply and quality of life for an impoverished region.

Health & Welfare
Aquaculture genomics: Progress in identifying species genetics continues
The continued application of genome research to aquaculture will provide unprecedented accuracy for genetic selection of performance and production traits.

Health & Welfare
Effect of dietary salt on Nile tilapia
Study examines impact of an increase in dietary NaCl on nutritional and other physiological processes, and effects on gut microbiome in Nile tilapia.

Health & Welfare
GIFT tilapia show greater FCR, growth potential than red tilapia
In a study of the effects of interaction between genotype and dietary protein levels on fish performance, GIFT tilapia had greater growth than red hybrids fed diets with equal or higher protein levels.



