Study develops, successfully tests a new L. salmonis-specific DNA-based assay for detection, quantification from seawater samples

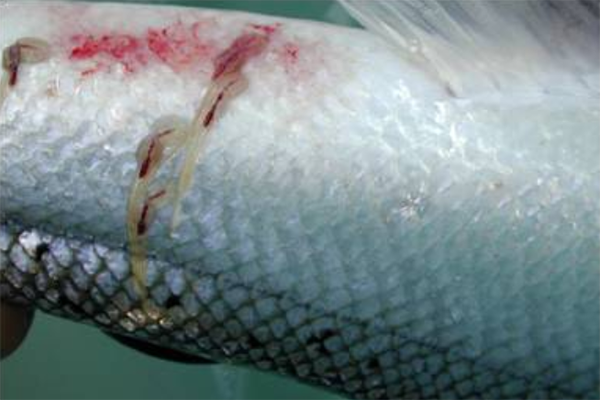
One of the largest challenges to the global salmon farming industry is infestations by various species of sea lice: primarily Lepeophtheirus salmonis and others. These parasitic copepods cause significant economic losses for the industry every year through decreased fish quality and reduced growth, treatment costs, secondary infections, stress and fish mortality. L. salmonis is commonly present in the natural environment both in the Pacific and Atlantic oceans and feeds on mucus, epidermal tissue, and blood on host salmonid species. Thus, the proliferation of salmon lice in intensive salmon farming does not only impose huge costs on the aquaculture industry itself but can also lead to significantly increased mortality for other salmonid species.
As L. salmonis can be a threat to the health and welfare of both farmed fish and wild fish, its density in the marine environment must be closely monitored, especially in areas close to aquaculture sites, and treatments must be done when needed. At present, monitoring of L. salmonis often involves manual inspection and counting of sea lice on farmed fish regularly. In several countries, this is set as a requirement by the government. The manual inspection of a certain number of fish per pen every week or every second week is a labor-intensive and costly approach, and also imposes significant handling stress for the fish.
Clearly, there is a need for a more cost-efficient, accurate, and less intrusive method for monitoring this species at any life stages to identify farm localities with high salmon lice density, and thereby high infestation pressure, at an early stage. Analysis of environmental DNA (eDNA) is a promising method for rapid and non-disruptive species detection in the aquatic environment and might therefore be used as a management tool to quickly assess the L. salmonis parasite load in the marine environment. eDNA is DNA extracted directly from an environmental sample without any physical collection or visual signs of the biological source material and hence eDNA can originate from cells shed into the environment from larger organisms.
This article – summarized from the original publication [Krolicka, A. et al. 2022. Sea lice (Lepeophtherius salmonis) detection and quantification around aquaculture installations using environmental DNA. PLoS ONE 17(9): e0274736] – reports on a study to design, validate and evaluate a new qPCR assay for detection and quantification of L. salmonis eDNA in the marine environment.
Study setup
In our study, we developed and tested a new L. salmonis-specific DNA-based assay (qPCR assay) for detection and quantification from seawater samples compatible with the Environmental Sample Processor (ESP) for autonomous water sample analysis of gene targets. For detailed information on the design, implementation and validation of assays; sample preparation and analytical work; and statistical analyses, refer to the original publication.
Results and discussion
Salmon lice is one of the most significant parasite threats to salmonid production in aquaculture and one of the greatest limiting production factors due to associated mortalities, as well as expenses related with the extensive and frequent treatments required. The issue has prompted the search for alternative adequate monitoring methods, ideally providing results in real time to be able to immediately introduce suitable actions to prevent large-scale salmon lice invasion and infection. In a recent study, none of available methods passed the comparative test of salmon louse enumeration in plankton samples. This suggests that a compromise among performance with precision, time used on the analysis and the costs must be made when choosing a method.
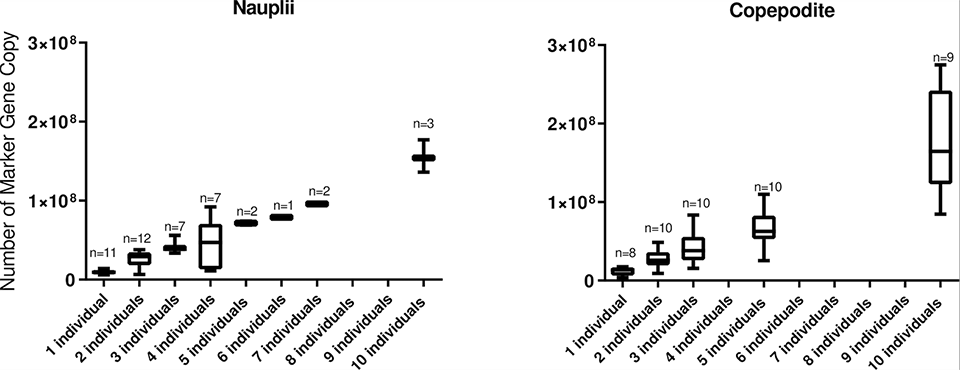
We demonstrated the specificity of the L. salmonis qPCR assay through in-silico [computer simulations] DNA analyses covering sequences of different L. salmonis isolates. Seawater was spiked with known numbers of nauplii and copepodite free-swimming (planktonic) stages of L. salmonis to investigate the relationship with the number of marker gene copies (MGC).
Field samples collected at different times of the year in the vicinity of a salmon production farm in Western Norway were analyzed for L. salmonis detection and quantification. The assay specificity was high and a high correlation between MGC and planktonic stages of L. salmonis was established in the laboratory conditions. In the field, L. salmonis DNA was consequently detected, but with MGC numbers below that expected for one copepodite or nauplii.
We detected L. salmonis eDNA at all four sampled locations around the salmon farm and from the three different sampled depth of 1, 5- and 10-meters. There were significant differences in the estimated L. salmonis MGC number at individual depths in September 2020, but not in May 2020. This possibly relates to different salinity, light and/or temperature conditions in these two months. In September, the light penetration to deeper layers of seawater was weaker than in May. The more homogeneous light and temperature conditions in May could have resulted in a more homogeneous distribution of L. salmonis and their eDNA.
The higher sensitivity and specificity of one of our tested assays (the SL2 qPCR assay) to L. salmonis can be a valuable tool for aquaculture monitoring. We obtained similar efficiency, but slightly lower correlation coefficients for both assays when compared to the other testing techniques. Both assays we tested, including the newly developed SL2 assays, are compatible with the ESP technology, which makes it possible in the future to investigate the potential for autonomous, on-site qPCR-based monitoring of L. salmonis.
Furthermore, even though the L. salmonis qPCR assay was only validated in the laboratory on L. salmonis obtained from one population of L. salmonis from the Atlantic Ocean, in silico analysis strongly indicates that the new assay should work for L. salmonis populations from both the Pacific and Atlantic Oceans. Recent studies have demonstrated weak population genetic differentiation among L. salmonis sampled not only from geographically distinct regions but also between the Pacific and Atlantic Oceans, suggesting a very high level of gene flow due to its high dispersal potential either passively by ocean currents or while attached to its highly migratory hosts.
Compared to the traditional methods, which are based on manually counting adult stages on the fish, an eDNA approach potentially has several advantages; 1) the results are delivered fast and efficiently; 2) it can reduce the time and costs associated with the monitoring as the method does not require host collection and manual counting; 3) it can detect a variety of stages of L. salmonis, including the first free-swimming stages, allowing for more representative measurements of the abundance and a better method to map the spread of infection in time and space; 4) it can cover several depths and seasons, and thereby produce more comprehensive biological data for L. salmonis abundance; 5) the method is non-invasive for the fish and thus does not affect fish welfare.
Furthermore, the qPCR assay was developed with the thought to be fully compatible with automation on the ESP. This instrument enables autonomous on-site sampling, filtration and DNA analysis of seawater samples, and further real-time streaming of results.
Using such a device would enable the farmers to obtain autonomous, frequent and rapid data on the presence of changes in the abundance of early stages of L. salmonis in the seawater. This would enable a rapid counteraction to mitigate the potential infection risk for the farmed fish. Should the ESP indicate a disease-free status; no actions are needed from the fish farmers. However, when the ESP reveals an increase in abundance (a problem), fish farmers will be able to take more immediate action to prevent an outbreak before fish get infected. This will most likely result in fewer fish being infected and less treatment needed, which could be beneficial in terms of fish welfare, economy, and environment impact.
Another advantage of using an ESP is that several qPCR assays can be included in one device, allowing for the detection and quantification of a range of target infectious or pathogenic species simultaneously. However, to use the eDNA approach for multiple diseases in aquaculture requires further development of protocols prior to the implementation into robotic devices such as the ESP. For instance, better methods for concentrating eDNA and size fractionation depending on sizes would increase the chances of the capture and detection of disease causative agents.
One challenge with using the eDNA approach to monitor L. salmonis is to establish how eDNA copy number relates to biomass. Compared to unicellular species, absolute individual-level quantification is complicated in animals where biomass, instead of count data, is likely to show a better correlation with DNA quantity. Furthermore, there is still some uncertainty related to the degradation rate and the dispersal rate of eDNA in aquatic systems. Several studies have suggested that eDNA can be used quantitatively, but for relative rather than absolute quantification. Experiments have shown a rapid degradation of eDNA in freshwater, and fewer studies have been performed on the persistence of eDNA in seawater.
Transport of eDNA within ecosystems could be a challenge in flowing waters and even more in marine environments. Nevertheless, the degradation of eDNA in aquatic systems has been found to occur at a scale of days or weeks, rendering long-distance dispersal unlikely. This is particularly important in large open systems such as oceans, where sea currents could potentially transport eDNA over large distances. However, there might also be species-specific differences in DNA persistence and this needs to be further investigated for L. salmonis eDNA.
Overall, the results of our study demonstrate the great potential for applying eDNA towards current challenges in aquaculture and gives promise for faster and less time-consuming manner of early detection of incoming threats.
Perspectives
We developed and analyzed a novel species-specific qPCR assay, which targets eDNA from the salmon fish parasite L. salmonis, and which is compatible with the ESP. The results can indicate that the SL2 qPCR assay can be used for reliable detection and quantification of L. salmonis eDNA in the water column. Thus, a DNA-based monitoring method that would not require host collection and manual counting, represents a relatively simple, non-intrusive and cost-effective alternative for monitoring of L. salmonis in the field and providing rapid notifications of potential infections to the farmers without causing welfare challenges for the fish.
The results from this study exemplify the usefulness of the eDNA approach and its potential as an alternative to standard monitoring practices. Nevertheless, calibration needs to be established to transform eDNA sea lice gene copy numbers into a sea lice individual count. To follow up this research and its application to aquaculture monitoring, there is a need to confirm the observations made herein with a larger data set and over a longer period.
Generally, if eDNA is to become a supplementary or alternative approach in sea lice assessment and monitoring for salmon farming, several challenges need to be investigated further. A promising avenue is also the implementation of such approach for real-time automation of sea lice detection such as with the ESP instrument. Our research offers very promising perspectives for the application of eDNA to aquaculture challenges.
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Author
-
Dr. Thierry Baussant
Corresponding author
Norwegian Research Centre AS (NORCE), Stavanger, Norway[111,110,46,104,99,114,97,101,115,101,114,101,99,114,111,110,64,97,98,104,116]
Related Posts

Health & Welfare
Barcoding, nucleic acid sequencing are powerful resources for aquaculture
DNA barcoding and nucleic acid sequencing technologies are important tools to build and maintain an identification library of aquacultured and other aquatic species that is accessible online for the scientific, commercial and regulatory communities.

Health & Welfare
New paradigms to help solve the global aquaculture disease crisis
Authors argue that interventions – like automated detection of pathogens and remote sensing applications – can help mitigate aquaculture's disease crisis.

Health & Welfare
Predicting parasite outbreaks in fish farms through environmental DNA
This article presents the results of a study using environmental DNA methodology for the early detection and quantification of various pathogens in farmed barramundi, where the technique showed to be a powerful and novel tool to predict fish mortalities.

Intelligence
DNA barcoding applications in aquaculture
DNA barcoding is an exciting tool for the identification of fish. Even non-taxonomists can classify organisms through the molecular taxonomy technique.



