Development of elite Asian sea bass lines using cutting-edge tools of genomics

The Asian sea bass or barramundi (Lates calcarifer) is a large-bodied, tropical marine teleost (bony fish), native to the Indo-Pacific region. Its farming originally developed in Southeast Asia, Australia and Papua New Guinea. Besides being one of the most popular food fish species for these regions, the Asian sea bass also entered the seafood markets of Europe and the United States at the end of the last century. Currently, most of its commercial production is primarily done by small family-owned and mid-sized farms, using brooders collected either from the sea or from unknown sources.
This article – adapted and summarized from the original publication (Orbán L. et al. 2021. Toward Genome-Based Selection in Asian Sea bass: What Can We Learn from Other Food Fishes and Farm Animals? Front. Genet. 12:506754) – reports on a recent review describing the current status of the process that was initiated nearly 15 years ago for the development of elite Asian sea bass lines using cutting-edge tools of genomics, discusses selection strategies used for improving the Asian sea bass lines and genomic resources and platforms generated for their use for the development of elite lines and for advanced studies of the biology species, and provides some practical advice for additional improvements in Asian sea bass selection and in aquatic selection programs in general.
Potential improvements for selection
Many important observations have been made by other researchers and by us over those decades when the Asian sea bass has been commercially aquacultured. Following we present a few suggestions for potential future improvements based on these observations.
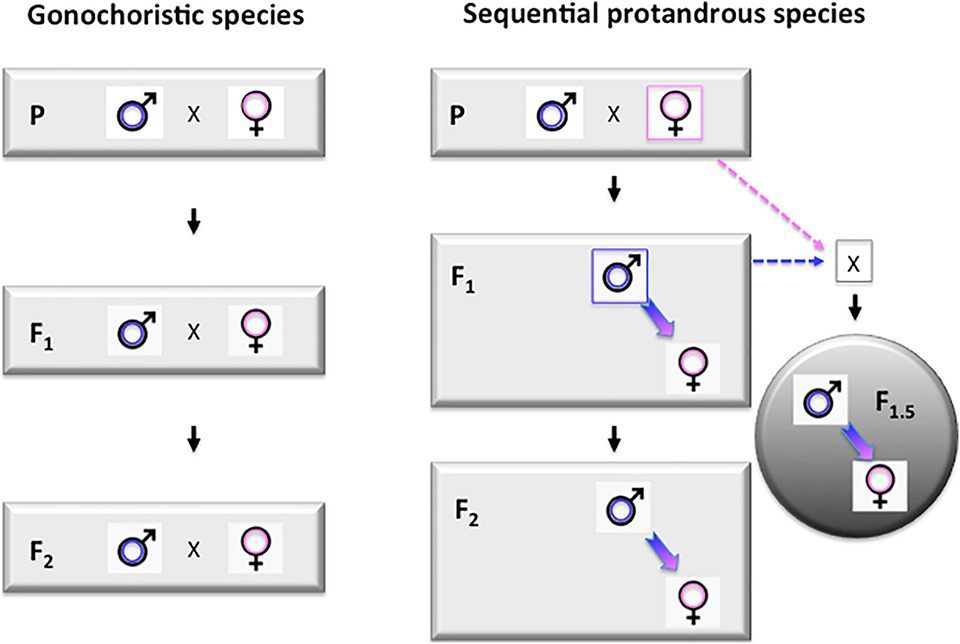
Due the long generation time (three to four years) of Asian sea bass, the selection process is rather slow. However, the sequential hermaphroditic [when an individual changes its sex at some point in its life; a sequential hermaphrodite produces eggs (female gametes) and sperm (male gametes) at different stages in life] nature of the species offers an interesting possibility to speed up the process.
The essence of this approach is based on the formation of so-called “intermediate generations,” as proposed earlier by other authors. Once the biggest individuals from the offspring groups of founding brooders reach a suitable size (around 50 cm standard length for individuals grown in seawater under tropical conditions), they start maturing as males. When a few of these young males reach maturity, they can be back-crossed with the founding females to produce a new generation.
We propose to call this the F1.5 generation, as they are F2s on the paternal side, but only F1s on the maternal side (Fig. 1). The performance of such F1.5 individuals is expected to be lower than the future F2s to be produced by the same F1 males crossed by future mature F1 females, but higher than the F1s.
During the selection process, we have occasionally formed breeding groups that contained a few siblings and half-sibs. Among the offspring of some these groups we have observed unwanted phenotypic signs – likely due to inbreeding effects – including loss of dorsal fins and other effects. These could be typically eliminated by genotyping the affected individuals, matching them to their parents and removing one or both of the carriers from the spawning group.
We have shown earlier that Asian sea basses are not uniform across the whole geographic area occupied by them. The three different types (Indian, South-East Asian and Australian) show substantial differences at the level of genome and phenotype as well. While natural populations might meet and potentially interbreed at bordering geographic areas, frequent transports of fertilized eggs or larvae between distant locations and even continents create additional opportunities for hybridizations within the species, but across these geographical varieties.
As the existing genomic platforms would make detailed analyses of potential connections between commercial stocks and natural populations around commercial facilities possible, we propose that such studies should be initiated to collect information about the frequency and extent of such hybridization events and their potential consequences.
Some considerations for food fish selection programs
Although the first fish species were domesticated thousands of years ago and have been selected since then based on their phenotypes, advanced selection programs supported by genetic/genomic platforms of food fishes have started much later than those of terrestrial farm animals. This delay offers certain opportunities, as researchers and farmers working on fish can utilize knowledge and approaches developed for those more advanced systems. The so-called “advantage of the follower” will allow aquatic selection programs for accelerated generation and rapid introduction of genomic platforms by following the examples of poultry, swine and other farm animals.
According to a recent survey, there are over three dozen selective breeding programs for the six most important food fish species in Europe, including the Atlantic salmon, rainbow trout, European sea bass and common carp. There are several other selection programs worldwide, mostly in China, India, Southeast Asia, Australia, New Zealand and the United States. The estimated number of species cultured in these regions is substantially higher than in Europe.
Examples include the GIFT project on Nile tilapia that supplied Southeast Asia with elite offspring with improved growth rate and survival, a 15-year selection program in striped catfish resulting in a 13.4 percent increase in growth rate, genomic selection using extreme phenotypes for increased weight in large yellow croaker and genomic selection to increase the resistance of Japanese flounder against a bacterial pathogen. For comprehensive reviews on aquatic selection programs see Lind et al., Gjedrem and Rye and Houston et al.
What are the main advantages and disadvantages of selection programs in fishes? Fishes are extremely prolific species with thousands or even millions of offsprings per family. In addition, the generation time of most food fish species tends to be shorter than for large-bodied mammals, so the initial goals of selection can be achieved earlier. The high number of offsprings allows for very high selection intensity (up to 2.51 in Nile tilapia and 1.75 in channel catfish), possibly higher than that for poultry. As the initial costs of keeping and growing fish larvae and juveniles are typically lower than for poultry, most tools and selection procedures that are applicable during the early developmental phases can be used to test much larger offspring populations in fishes.
On the other hand, the price of an offspring individual for most cultured fish species is typically very low, comparable to that of poultry. This prevents large-scale use of expensive genomic technologies (e.g., whole genome re-sequencing) and forces researchers to consider more cost-effective alternatives. Another important factor to consider is the early growth rate, as juveniles need to reach a certain size before they can be tagged physically or with RFID tags for subsequent re-identification following genotyping.
The ultimate genomic tool is a sequenced and assembled genome. Although a reference genome is not an essential requirement for genomic selection, the availability of information at such a high-resolution might exert obvious beneficial effects on such projects. The list of fish species with sequenced and assembled genome is rapidly expanding despite the fact that they are more difficult to deal with due to their increased genome complexity.
Currently the number of assembled teleost (bony fishes) genomes stands around 100 and it will likely double within a year. Moreover, there are several large-scale efforts that target the genome sequencing and assembly of additional hundreds or even tens of thousands of vertebrates, among them many fishes [for example, see the Vertebrate Genomes Project).
Perspectives
Aquatic selection programs have entered the era when their dependence on genomic tools and platforms will drastically increase. By following the footsteps of those working with terrestrial farm animals, researchers have been developing genomic toolboxes for an increasing number of food fish species used in aquaculture production.
This review describes the history of the selection program performed on Asian sea bass in Singapore over the last 15 years. It covers the protocols applied at the farm, the full genomic toolbox developed for the species and their application in different studies. As this might be the first selection program performed on a tropical, sex changing predator, some of the unique aspects stemming from these features are also described. A few considerations for aquatic selection programs are also provided in the hope that they will be useful for those who are in the planning phase of such projects.
We hope that our review will be useful not only for researchers and farmers working with Asian sea bass, but potentially for those who face similar decisions in the work with other, lesser-known tropical food fishes as well.
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Authors
-
Dr. László Orbán
Corresponding author
Reproductive Genomics Group, Temasek Life Sciences Laboratory, Singapore, Singapore; and Frontline Fish Genomics Research Group, Department of Applied Fish Biology, Institute of Aquaculture and Environmental Safety, Hungarian University of Agriculture and Life Sciences, Keszthely, Hungary[117,104,46,101,116,97,109,45,105,110,117,64,111,108,122,115,97,108,46,110,97,98,114,111]
-
Dr. Xueyan Shen
Reproductive Genomics Group, Temasek Life Sciences Laboratory, Singapore, Singapore; and Tropical Futures Institute, James Cook University, Singapore, Singapore
-
Dr. Norman Phua
Reproductive Genomics Group, Temasek Life Sciences Laboratory, Singapore, Singapore
-
Dr. László Varga
Institute of Genetics and Biotechnology, Hungarian University of Agriculture and Life Sciences, Gödöllõ, Hungary; and Institute for Farm Animal Gene Conservation, National Centre for Biodiversity and Gene Conservation, Gödöllõ, Hungary
Tagged With
Related Posts

Intelligence
Barramundi culture in Australia
From recirculation units, earthen ponds and sea pens in Australia, barramundi are spreading to other continents due to the advent of sophisticated modular culture systems.

Intelligence
Full speed ahead for Florida pompano RAS farm
While many attempts to farm Florida pompano have fallen short, Joe Cardenas is undaunted, confident that recirculating aquaculture systems are the key.

Responsibility
Australis Aquaculture wins prestigious climate innovation award
Australis Aquaculture received the U.S. Secretary of State’s 2021 Award for Corporate Excellence in the Climate Innovation category for advancing climate-smart ocean farming of barramundi in Vietnam.

Innovation & Investment
Israeli firm’s ‘plug-and-play’ RAS solution attracts investment
BioFishency's two products, the Single Pass BioFilter and the Mini-RAS, each aim to improve the survival, growth and reproduction rates of fish, which increases profitability and sustainability for the farmer.



