A total of 34 species from 14 families were identified at the species level from 164 sequences

The advent of the genetic barcode has improved the capacity to resolve the misnaming of species, to detect seafood and product fraud, to minimize the trafficking of endangered and CITES-protected (International Convention for Endangered Species Trade) species, and to reduce illegal, unregulated and unreported fishing (IUU). In addition, barcodes allow for the identification and discovery of new species, separation of cryptic species, species complexes, stock characterization, detection of molecular hybrids and understanding migrations and connectivity patterns.
In fishes, the use of a barcode began in 2003; and by 2009, more than 5,000 fish species were characterized by this method. Considering that the Osteichthyes or bony fishes are the most diverse vertebrate taxa, with more than 28,000 species reported, it is evident that scientists still have some important work to carry out. The implementation of a traceability system in Latin America for fish and fishery products is currently in an early development phase or yet to be implemented. Only countries such as Peru, Chile and Ecuador have implemented a traceability model for any fishery.
The fish fauna of Panama is highly diverse and includes 1,412 species, of which 1,240 are marine species. Approximately, 814 of these species have been reported in the Pacific coast of Panama and this represents 60 percent of the fish diversity for the country, making it the most diverse vertebrate taxa. In a recent review, a total of 223 commercial fish species were reported for the Pacific side of Panama, corresponding to 183 bony and 70 cartilaginous fish. These species are locally commercialized or exported as fish products (fillets, fins and whole fish).
This article – summarized from the original publication (Díaz-Ferguson, E. et al. 2023. Building a Teleost Fish Traceability Program Based on Genetic Data from Pacific Panama Fish Markets. Animals 2023, 13(14), 2272) – reports on research to provide a DNA barcode list of Pacific Panama commercial fish and the first haplotype (group of alleles – matching genes from biological parents – in an organism that are inherited together from a single parent) candidates for molecular traceability as well as the first values of genetic diversity, connectivity and demographic history for the most common species collected in the largest fishery areas of Pacific Panama (Gulf of Panama and Gulf of Chiriquí). This is also a pioneering study, providing potential markers for the most common species.
Study setup
Fish tissue samples from 203 adult individuals were collected in the main ports and markets of the Pacific coast of Panama. Main fishery landing ports of the Pacific coast of Panama (Coquira, Pedregal and Mutis) and the biggest fish gathering centers of the Pacific side of the Country (Mercado del Marisco-San Felipe Market, Mercado de Anton, Mercado de Santiago and Isla Verde-Golfo de Montijo) were visited (Fig. 1).
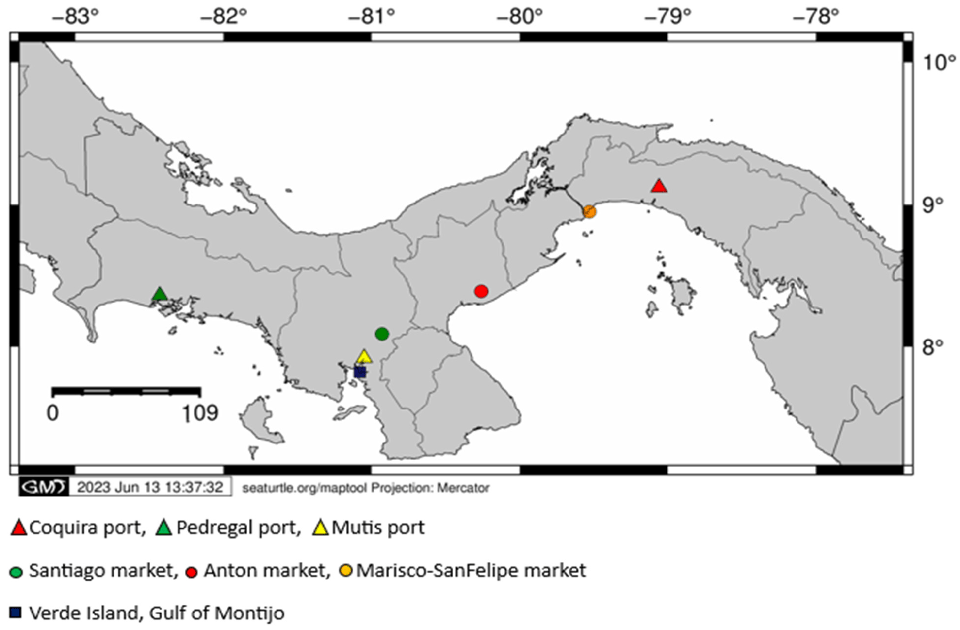
Additionally, red snapper samples from the Pixvae area, located within the buffer zone of Coiba Island National Park (largest marine park in Central America and UNESCO Natural Heritage Site), were also analyzed. At each collection site, fin clips and muscle tissue of the main fish families reported as commercial for the Pacific Panama were collected, labelled and stored for analysis. Molecular identification based on a cytochrome oxidase I gene segment of all species was verified by GenBank® [the U.S. Department of Health and Human Services’ National Institutes of Health (NIH) genetic sequence database, an annotated collection of all publicly available DNA] reference sequences.
For detailed information on the sample collection procedures; DNA extraction and PCR amplification, and sequence analysis, refer to the original publication.
Results and discussion
Fish tissue samples from 203 adult individuals were collected in the main ports and markets of the Pacific coast of Panama. Molecular identification based on a cytochrome oxidase I gene segment of all species was verified by GenBank® reference sequences. A total of 34 species from 14 families (Ariidae, Caranjidae, Centropomidae, Gerreidae, Haemulidae, Lobotidae, Lutjanidae, Malacanthidae, Mugilidae, Scianidae, Scombridae, Serranidae, Sphyraenidae, Stromateidae) were identified at the species level from 164 sequences.
This is the first molecular identification study and the largest number of DNA sequences deposited in Genebank® for marine commercial teleost species from the Pacific coast of Panama in a single publication, including the main fishery areas. No similar data sets have been previously reported for the Eastern Tropical Pacific.
This research is also a pioneer study because we provide the first molecular list of commercial bony fishes collected and commercialized along the main fishery areas of the country including different points of the commercial chain, i.e., landing sites and fish markets. The total number of species and families obtained in this study represents a high percentage of the total commercial species already reported for Panama’s Pacific coast using traditional taxonomic methods.
Distinct stocks of Atlantic cod face different climate change challenges
Some other species listed in previous taxonomic and fishery studies for Panama’s Pacific coast were not collected in the present study, including some drums, tunas and other species. One possible explanation is that nearly all of our samples were collected during visits to landing sites, markets and gathering sites with the exception of red snapper samples from Coiba National Park, collected from artisanal fishermen. The absence of large pelagic species such as tunas, dolphin fish and bonito in our analyzed landing sites and fishery markets were due to the direct commercialization of these species from the industrial fishery sector to international boats and external processing plants.
Another finding was the existence of species reported in the Caribbean side of the country such as Mycroperca xenarcha, Paralonchurus dumerilli and Lobotes surinamensis in gathering sites and public markets of the Pacific side of the country as evidence for local commercialization and traceability of Caribbean species into bigger Pacific markets with a higher consumer demand.
Understanding genetic diversity patterns can help us to understand the sustainability of fisheries and fitness. Higher values of this parameter indicate which species are likely to adapt to future disturbance. Genetic diversity is also a measure of reproduction strategy, fecundity, effective population size and life history variation based on habitat type and period of larval development. Genetic diversity values make it possible to establish comparisons of these parameters for species occupying different habitats or having different life histories. Despite its importance, genetic diversity data are limited and are regionally oriented mainly epipelagic species, i.e., tunas and jacks.
The study of population genetics, demographic history and genetic connectivity is an important tool for fishery management and understanding population structure, reproduction patterns, migration and passive larval dispersal as well as determining selection pressures that allow scientists and decision makers to generate conservation strategies in fish exploited populations. We examined the connectivity patterns of three species: the spotted rose snapper (Lutjanus guttatus); the green jack (Caranx caballus); and the Pacific sierra (Scomberomorus sierra). Only L. guttatus showed significant differences among the three fishery areas (Gulf of Montijo, Gulf of Panama and Gulf of Chiriquí). However, elevated connectivity or lack of genetic differentiation among Gulf areas was evidenced among samples of S. sierra and C. caballus, specially between the Gulf of Chiriquí and the Gulf of Panama.
Molecular data from Panama commercial fish species is not available for most of the listed and reported fish species. Through this research we provide primers, probes and target amplicon regions for qPCR eDNA detection of five important commercial fish species (Lutjanus guttatus, Lutjanus argentiventris, Caranx caballus, Caranx caninus, Scomberomorus sierra). These primers and probes could be used in testing to avoid fraud in fish exports and the use of false tariff codes for exportation.
In addition, of the newly developed markers, new obtained haplotypes were deposited on Genebank for all analyzed species. This information will become a reference for fishery studies providing the origin and fishing area of these species. Additionally, fish mitochondrial haplotypes have been used for determining and tracking farm fish population origins as well as to introduce new variants into hatcheries to improve genetic variability, fitness and breeding programs.
Conservation and management of genetic diversity is key for sustainable fisheries. The results of our study provide important information and molecular tools for accurate identification of species, its traceability along the supply chain by providing species-specific tariff codes using a binomial species-specific nomenclature system, conducting species-specific biodiversity monitoring and fishery management.
Additionally, this information contributes to the future generation of a molecular database for accurate stock identification, genetic health programs, the establishment of population boundaries, connectivity patterns, understanding population patterns and effective management of population size. A better understanding of these features will provide accurate information for decision-makers and policy planners that contribute and support national and regional policies based on scientific data and create a balance between the power of the industrial fishery sector, local governments, and fishery managers.
Thus, by providing the first haplotypes and values of genetic diversity for some species such as L. guttatus, S. sierra and C. caballus collected in Panamanian waters, we form the molecular database that will be a step forward for the future establishment of a marine species and fishery product traceability program that will be crucial to achieving an ecosystem management approach to our fisheries, recovering overexploited stocks and reducing illegal, unreported and unregulated fisheries (IUU).
Perspectives
The results of this study represent the first attempt at providing a molecular database (GenBank® accession numbers) of Panamanian commercial fisheries, i.e., fish species and fishery products detected molecularly. This information is the first step toward the establishment of a future fish traceability program in the country and is supported by molecular data. These data will be key not only for the traceability of commercial species but also for future genetic stock assessment, the establishment of conservation units and sustainable fishery programs. In addition, having a strong traceability system will strengthen the Hazard Analysis and Critical Control Point (HACCP) system of the country and minimize the impact of IUU in the transparency of our fisheries.
This research also contributes with the first species-specific primers and probes designed in silico (an experiment performed on computer or via computer simulation) for environmental DNA (eDNA) detection and traceability of some of the most common commercial fish species reported in Panama waters. The production of molecular markers for eDNA detection of commercial species will be the base for a future molecular traceability pilot program in Panama with the potential to become a model for Latin America and the Greater Caribbean region. In the future, similar efforts should be considered for sharks, rays and billfishes since several populations are currently endangered, banned or have CITES status.
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Author
-
Carlos Ramos-Delgado, Ph.D.
Corresponding author
Coiba Scientific Station (COIBA AIP), Gustavo Lara Street, Bld. 145B, City of Knowledge, Clayton, Panama City 0843-01853, Panama; and Faculty of Natural and Exact Sciences, Department of Genetics and Molecular Biology, University of Panama, Panama City 0824-3366, Panama[97,112,46,99,97,46,112,117,64,100,115,111,109,97,114,46,115,111,108,114,97,99]
Tagged With
Related Posts

Innovation & Investment
‘We have 15 years to fix this planet’: Blue Food Innovation Summit explores potential of restorative aquaculture – and the challenges to scaling
At the Blue Food Innovation Summit, discussions centered on restorative aquaculture’s potential and financing the ocean’s capabilities.

Health & Welfare
Barcoding, nucleic acid sequencing are powerful resources for aquaculture
DNA barcoding and nucleic acid sequencing technologies are important tools to build and maintain an identification library of aquacultured and other aquatic species that is accessible online for the scientific, commercial and regulatory communities.

Intelligence
Can a ‘chemical fingerprint’ deter seafood fraud?
Because paper trails aren’t perfect, some food producers are going beyond written or digital records to prove their products’ authenticity and prevent economic fraud. Is farmed seafood a perfect fit for forensic-science traceability?

Intelligence
In the fight against seafood fraud, the technology behind trace element fingerprinting is maturing
Analysis of trace chemical elements can reveal where farmed seafood comes from. Standardization of traceability tools and techniques is improving the process.



