Successful genomic selection program for next-generation shrimp broodstock in Ecuador uses genomic data to improve survival and growth

Ecuador increased its farmed shrimp production five-fold in the last 10 years, reaching over 1 million metric tons in 2021 (Fig. 1). This was achieved without specific pathogen free (SPF) shrimp, and without foreign broodstock purchases, genomic selection (GS) or fancy marketing claims, with just silent and anonymous hard work, devoted and constantly improved management skills, and the implementation of functional technological solutions such as efficient multi-phase grow-out systems, automated feeders, improved feeds, more effective water quality management and bottom soil bioremediation approaches, and effective local genetic programs.
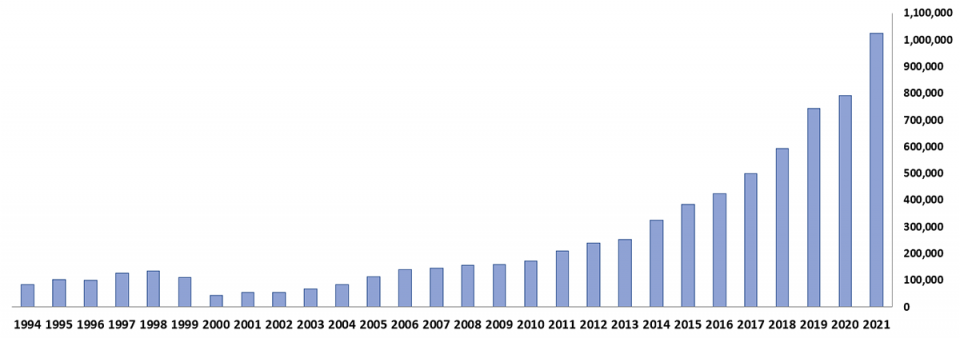
It is an accomplishment unmatched by any other shrimp producing country in the world during this period. In the last eight years, the best shrimp farms in Ecuador have more than doubled their shrimp growth rates, from under 1 grams/week at a stocking density of 10 shrimp per square meter to over 2 grams/week at 15–25 shrimp per square meter; doubled the productivity per hectare, from 3,500 pounds per hectare to 7,000 pounds/ha; considerably shortened the grow-out cycles from 180–200 days to 100–120 days, which represents one additional grow-out cycle per year; and significantly reduced the feed conversion rates from ~1.9 to ~1.4.
On the genetics front, effective local breeding programs have been very successful with respect to growth rate improvements and inbreeding management, but less successful in determining predictable improvements in the survival rates whose performances have been more impacted by the vagaries of year and management effects. Inspired and propelled by its success in the improvement of the growth rates of their genetic stocks, Texcumar, the leading maturation and genetic program in Ecuador, with a daily production of 400 to 500 million nauplii (compared to 100 million per day 10 years ago), decided in a pre-pandemic setting to launch a genomic selection (GS) program with the objective of evaluating a new state-of-the-art genetic tool which might enable us predictable and sustained genetic responses for farm survival rates, the missing component in our otherwise very successful mass selection program.
Genomic selection
Throughout the last decades, with the development of next-generation sequencing technologies [which can very quickly sequence or decipher the genetic make-up of an organism] and the identification of large numbers of DNA molecular markers of the Single Nucleotide Polymorphism, or SNPs [a single base pair variation in the DNA of an organism] type, detailed genome sequencing and low-cost assessment of SNPs with full genome coverage became viable for virtually any species. The concept of genomic selection (GS) was first introduced by Meuwissen, et al. and then later gradually implemented and expanded into a condition of universal acceptance as the last frontier and most powerful approach in genetic improvement.

Genomic selection can be defined as a strategy of genetic evaluation and selection based on a very large number of genetic markers (over 10,000) which enable a full genomic coverage, such that all genetic loci [singular locus; a fixed and specific position on a chromosome] impacting any particular trait of interest will be associated through linkage disequilibrium [a sensitive indicator of the population genetic forces that structure a genome [all genetic information of an organism] with at least one of the genetic markers.
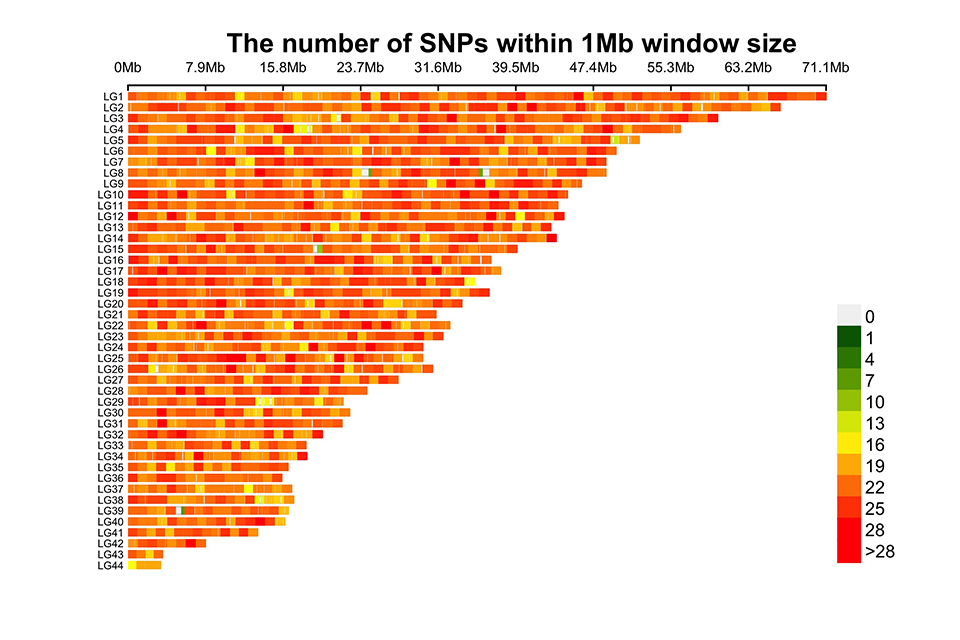
This concept is illustrated in Fig. 2, which represents the distribution – in the 44 chromosomes of the Pacific white shrimp (Litopenaeus vannamei) – of the 50,000 SNP markers composing the genomic chip of the Pacific white shrimp (Litopenaeus vannamei) developed by the Center for Aquatic Technologies (CAT) in conjunction with Neogen in the United States. This genomic tool is already commercially available for different types of applications and genomic selection strategies in L. vannamei.
In the footsteps of previous genetic improvement models – such as Mass Selection (based on multiple spawns), Family Selection (based on individual spawns under different types of identification systems), and the less generalized Marker-Assisted Selection (capitalizing on only a few individual genic effects of larger magnitude) – genomic selection has been introduced as the most advanced and powerful avenue for genetic improvement currently available, capable of leveraging information on and of directly measuring a very large percentage of all individual gene effects impacting any particular trait of interest, thus leading to greater accuracy in the selection of broodstock and powering faster genetic gain rates for the aggregate economic net merit of any particular species than any of the former genetic improvement approaches.
This concept has rapidly disseminated, and genomic selection has now become the backbone of dairy genetic improvement, and also with significant contributions in other species, such as swine, poultry, some plant species and beef cattle.
In aquaculture, the concept is also gaining momentum, being most advanced and widely utilized in Atlantic salmon breeding, but also being considered in other species, even if in more incipient development stages as compared to those of the terrestrial species. For L. vannamei shrimp, Australia and Indonesia have generated the first advances, although in a more preliminary and academic context. More recently, Lillehammer, et al., Robinson, et al. and Hennig, et al. (2021. Developing SPR Lines from SPF populations using Genomic Selection. Book of Abstracts Congreso Mundial de Acuacultura AquaExpo Guayaquil, Session 4 Genetic Improvement:33.), have produced and reported solid work to develop lines resistant to the White Spot Virus (WSSV) based on genomic selection.
Genomic selection for resistance to White Spot Syndrome Virus in Pacific white shrimp
Then in 2020, Texcumar in Ecuador launched a commercial GS program, and in our opinion and to our knowledge, this is the first genomic selection project that fully links and integrates with the commercial pipeline of a large-scale shrimp genetic improvement program, with controls in place to measure the impact versus standard commercial lines.

The genetic improvement advantages and benefits that have been attributed to and associated with the implementation of genomic selection include:
- Increases in the accuracy and reliability of all genetic parameters and of breeding values, especially for low heritability traits such as survival.

This project included a total of 12,000 pleopod collections, with 12,000 weights measured and 12,000 eye tags inserted over a two-week period – and then collating all this data with the rigor required for a genomic selection project. Photo by Texcumar. - Greater flexibility in the implementation of effective genetic schemes, enabling phenotypic [observable physical characteristics] data collection in commercial environments and in specific disease challenges as additional selection criteria.
- Greater, and more precise and effective inbreeding [offspring from the breeding of organisms that are closely related genetically] management.
- Implementation of selection schemes at much earlier stages in the lifecycle, such as postlarvae (PLs) and juveniles, therefore reducing costs and generation intervals.
- More effective selection schemes for traits whose evaluation requires sacrifice of the animals, such as final product sensory and composition attributes (meat quality, omega 3 composition).
- All the above leading to considerable faster rates of genetic gain for the aggregate economic net merit of any given species.
With the application of genomic selection, additional genetic gain rates of 15 to 120 percent per year have been documented for different species (see references cited earlier).
Implementation of genomic selection in Ecuador
Our objective was to select a group of animals as broodstock for the next generation using genomic data to improve primarily survival, but also growth rates.
The outline of the plan:

- Place 80 fertilized shrimp females in spawning tanks. After the nauplii hatch, distribute them equally among four larval rearing tanks (LRT).
- Collect samples of pleopods [abdominal appendages or legs] from the fertilized females and all potential mates. These animals were genotyped with the 50K high-density chip.
- At the end of larval rearing 1,500 PLs from each LRT were randomly collected for genotyping using a low-density (LD) chip to evaluate family representation before moving to nursery ponds.
- At the end of larval rearing the PLs from each LRT were distributed equally among four nursery ponds.
- At the end of the nursery phase, tissue samples from 3,000 juveniles per nursery pond were randomly collected and used to evaluate family representation at the end of the nursery phase with a LD chip.
- Each nursery pond was transferred to a grow-out pond.
- When the grow-out ponds were harvested, 4000 animals were selected at random from each grow-out pond and were transferred to maturation, and 3000 of these were eye-tagged, weighed and a pleopod was collected for genotyping using the high density 50K chip.
- Genomic-Estimated Breeding Values, GEBVs [genetic potential of the animals] were calculated for the 12,000 animals, and the 800 females and 600 males with the best genetic potential were selected for broodstock to produce the next generation.
Statistical analyses and genomic evaluations
Survival: The ponds were sampled at the end of each stage in the cycle and the relative survival of each family was determined by the change in family representation in the sample from stocking to harvest. This response is based on the proportion of individuals that belonged to one of the identified 73 families that were part of the study. A baseline for this proportion was considered at the initial sample, at the transfer to the grow-out tanks (from the nursery tanks), and then at the end of the experiment a second sampling was done at the harvest from the grow-out tanks. To ensure good estimates of survival, 3,000 individuals were sampled from each pond at harvest, for a total of 12,000 progeny. Statistical calculations of sample size indicated that this number (3,000 per pond assigned back to family) is sufficient to detect differences in family survival with reasonable accuracy.
Using mathematical models of Genomic-BLUP, GBLUP [the method of genomic best linear unbiased prediction uses genomic relationships to estimate the genetic merit of an individual] for survival (DA), the parent-offspring relation was calculated by measuring the genetic similarities throughout the genome. A Parental Model was used to estimate a breeding value for each of the parents used in the dataset. The model considered each of the ponds as a different environment, and a different genetic variance was estimated. A common additive genetic correlation, rA [equivalent to the correlation between breeding values] was obtained among all ponds, together with a single error variance.
As a second stage, the GEBVs for each of the parents obtained from this model were used to estimate the offspring genetic potential for all the other individuals with genomic information, by using the approach of conditional estimation based on a genomic relationship matrix with all 12,663 genotypes. Since each pond was analyzed separately, these models provide four survival GEBVs for every genotyped individual, allowing for specific Survival selection for each of the different environments.
For a detailed description of the base genetic model used, contact the corresponding author.
In this model, pond is the fixed effect of the ponds, and genotype is a random effect associated with the genomic matrix. This model considers a common correlation between ponds but with heterogeneous genetic and error variances considered for each of the ponds. Hence, this allows each pond to have different heritability and therefore different precision and contribution on the genetic signal.
Growth: A total of 12,663 animals were genotyped [defining differences in the genetic make-up, or genotype, of an individual] from the samples collected when the ponds were harvested. A weight genomic breeding value was calculated for all animals based on the phenotypic data and the genomic relationships. Assuming an approximate normal distribution for the weight response, a genomic prediction animal model was fitted for this data, based on the genotypic data. For detailed information on data processing, contact the corresponding author.
With the fitting of this model a genomic prediction, GP [tool that can predict an animal’s genetic merit based on scoring certain DNA markers] was obtained for all the individuals that were genotyped. All different ponds were considered a single environment in the context of the growth analyses. The GP values correspond to the overall mean + GEBVs.
Preliminary results and discussion
The results from fitting the multi-environment GBLUP model to DA (Survival) resulted in heritability estimates of 0.08 ± 0.13, 0.23 ± 0.18, 0.23 ± 0.19 and 0.46 ± 0.17 for the four different ponds/environments. The GxE [genotype-environment interaction] evidence gathered led us to group two of the ponds into a single environment, and thus consider only three different Survival environments overall.

The correlations between the growth and the survival genomic breeding values for each pond were low and not significantly different from zero. Thus, the genomic breeding values for growth and survival appear to be almost independent of each other. No disease outbreaks were reported during the grow-outs and under such no disease scenario a null correlation between growth and survival is a reasonable expectation now corroborated by the genomic findings. Regarding growth, the fitted genomic animal model (Bayes B approach) resulted in a heritability of 0.39, which is a reasonable estimate for a growth trait, when compared to other studies.
Three environment-specific survival lines were established (high, medium and low density), and our results indicate that we can expect an increase in survival ranging from 3 to 8 percent among the three different environments considered. This is the improvement in survival expected if crosses between the highest ranked animals occur and the progeny from these broodstock experience very similar environmental conditions. Emphasis was placed on selection for survival and on genetic variability management to establish a sound foundation for future gains.
These are preliminary results. Farm results from the first-generation offspring selected under GS protocols are now under full scrutiny and definitive conclusions will be the subject of future communications. We had much to learn during the process. Mass Selection is a fairly simple scheme to run efficiently. To jump from Mass Selection to Genomic Selection represented a major challenge in procedures and personnel skills. We were able to implement and complete this first large-scale GS project, but with a number of errors and missteps. And the costs incurred are high. To beat the incredible efficiency of Mass Selection for growth under such high costs is a difficult proposition even for GS. And survival remains a very thorny trait to improve even under GS. With the high costs involved, dilemmas ensue regarding: the appropriate balance between genetic variability management and immediate selection gains; and the appropriate consideration of GxE interactions and how many different environments we can and have to really consider.
Considering the missteps, the learning curve, and the costs, some of the initial results appear promising. We are certainly learning and thinking. The advantages of genomic level variability management have emerged as very interesting. And with joint projects involving several groups the costs can be diluted. The future is upon us every day, and this first GS project was for us above everything else a learning experience. And from what we have learned, with the lessons gained, it appears as if GS deserves some additional commitment and scrutiny. We should emphasize this was not an academic experimental project, but a serious and committed effort to integrate genomic selection fully and functionally into the commercial pipeline of our large-scale breeding program in Ecuador.

Conclusions
To our knowledge, ours has been the very first large-scale shrimp genomic selection project aiming to fully integrate the pipeline of a commercial shrimp breeding program of large dimension. In our opinion and to our knowledge, back in 2020 when it was launched, this was the first wide-scale application of genomic selection truly at commercial scale, with controls in place to measure the impact versus standard commercial lines.
We are glad and proud that Ecuador took the lead in this endeavor, affirming its role as the world’s largest shrimp producer and exporter in 2020 and 2021. We found the jump from mass selection to genomic selection to not be an easy proposition at commercial scale. But with all things considered, the first results on the ground appear promising and interesting enough to deserve a second look, most especially the advantages that were identified as conferred by the sophisticated genomic variability management now made possible by GS, and which are not amenable to the mass selection schemes.
Industry consolidation and other future trends that may take place may change the perspective on many of these issues. Genomic selection certainly appears more appealing than classic family selection, an idea and an approach difficult to justify when GS is now available. But we are still evaluating if under current scenarios GS delivers favorable cost-benefit ratios that could enable it to outplace or even realistically complement a well-oiled and very efficient low-cost mass selection machine. Above all, our results comprise a great learning experience and the lessons learned were very positive. Genomic selection in shrimp breeding may get in slowly and with a less than smooth path, but it is a concept that has probably arrived to stay.
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Authors
-
João L. Rocha, Ph.D.
Corresponding author
Texcumar S.A., San Pablo, Santa Elena, Ecuador[109,111,99,46,111,111,104,97,121,64,97,104,99,111,114,99,108,108,111,97,111,106]
-
John Buchanan, Ph.D.
Center for Aquatic Technologies, San Diego, California, USA
[109,111,99,46,114,101,116,110,101,99,104,99,101,116,97,117,113,97,64,110,97,110,97,104,99,117,98,106]
-
Salvador A. Gezan, Ph.D.
Center for Aquatic Technologies, San Diego, California, USA
-
Richard Towner, Ph.D.
Center for Aquatic Technologies, San Diego, California, USA
-
Jason Stannard
Center for Aquatic Technologies, San Diego, California, USA
-
Rafael Verduga
Texcumar S.A., San Pablo, Santa Elena, Ecuador
Tagged With
Related Posts

Health & Welfare
Genetic improvement in Brazil
Breeding efforts at Aquatec Genearch Aquacultura in Brazil have centered on specific pathogen-free white shrimp lines imported from the United States.

Health & Welfare
Shrimp genetic improvement in Ecuador
One of the largest shrimp maturation operations in Ecuador is working with five major shrimp producers to improve growth rates.

Health & Welfare
Within-family selection enhances shrimp growth
A study in Brazil found that the growth superiority brought about by within-family selection of tested shrimp transmitted to the next generation of animals.

Health & Welfare
Shrimp hybrids outperform parents in Brazil studies
In Brazil, shrimp hybrids – crosses of imported specific pathogen-free lines and a genetically improved and locally adapted line – outperform their parents.



