Results show improved survival under an experimental challenge test

Disease caused by the White Spot Syndrome Virus (WSSV) is a serious problem for the global shrimp farming industry, as it infects all major cultured shrimp species and is highly virulent, normally causing death in a few days. Aquaculture practices, mostly aimed at eliminating the causal agent, may reduce the economic risks associated with White Spot Disease, but elimination in open pond culture systems is often impossible.
The simultaneous improvement of growth and WSSV resistance with traditional selection is complicated by a negative genetic correlation between these traits. The reported genetic gain per generation for WSSV resistance ranges from 1.7 to 6.5 percent. Genomic selection [a concept based on using DNA information to obtain a direct genetic value of the animal] has been proposed as the most effective form of selection available for typical quantitative traits [observable traits determined jointly by many genes with small effects and the effect of the environment]. Genomic selection could be used to improve the genetic gain in WSSV resistance.
This article – adapted and summarized from the original publication [Lillehammer, M. et al. 2020. Genomic selection for white spot syndrome virus resistance in whiteleg shrimp boosts survival under an experimental challenge test. Sci Rep 10, 20571 (2020)] – reports on a study to determine the power and accuracy of genomic selection to improve WSSV resistance in Pacific white or whiteleg shrimp (Litopenaeus vannamei). Shrimp were experimentally challenged with WSSV and resistance was evaluated as dead or alive (DOA) 23 days after infestation.
Study setup
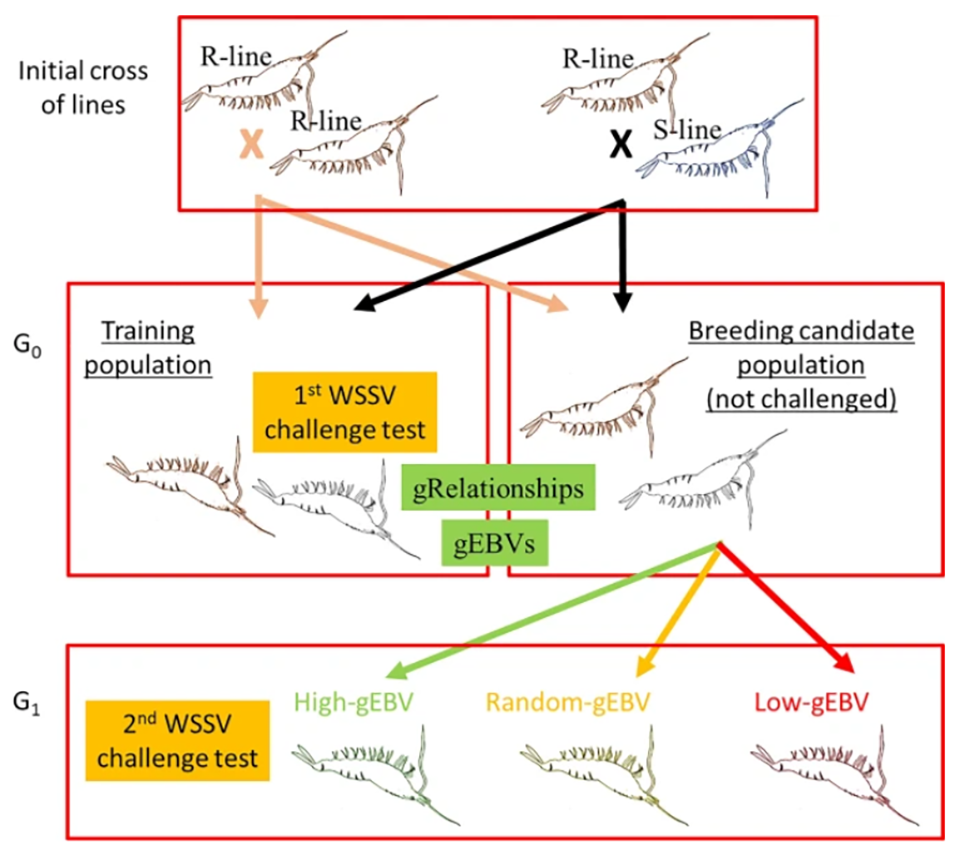
In our study, we used two populations of L. vannamei that had been selectively bred for several generations by Benchmark Genetics (formerly Ceniacua) in Colombia (Fig. 1). The first population was derived from a breeding operation on the Atlantic Coast of Colombia that commenced in 1997; this population has not been exposed to WSSV and is herein referred to as the susceptible line (S-line). Combined family and within-family selection of this population focused on fast growth, resistance to Taura Syndrome Virus, general pond survival and robustness for 16 generations under strict biosecurity protocols. The second population, herein referred to as the resistant line (R-line), was derived from a Pacific Ocean population in 2008, and it was mass selected with high intensity for survival in the presence of WSSV for seven generations.
These Atlantic and Pacific shrimp populations were maintained in isolation on their respective (Caribbean and Pacific) coasts of Colombia. All animals used in our study were considered naïve (uninfected with the virus). Animals from the S-line were PCR-tested every three months for all shrimp pathogens listed by the World Organisation for Animal Health (OIE) and were found to be negative for all listed pathogens for more than four years. Before the challenge test, a representative sample of the animals from the R-line was also PCR tested and found to be negative.
For detailed information on the experimental design and animal husbandry; WSSV challenge tests; tissue sampling and genotyping; and various genetic and statistical analyses, refer to the original publication.
Opinion: OIE, the World Organization for Animal Health, needs seafood industry’s input
Results and discussion
All shrimp in the challenge test were genotyped to determine differences in the genetic make-up of individuals for 18,643 single nucleotide polymorphisms, SNPs [a substitution of a single nucleotide (basic building blocks of nucleic acids, DNA and RNA,) at a specific position in the genome; SNPs can be used to pinpoint differences in susceptibility to a wide range of diseases]. Shrimp breeding candidates (G0) were ranked on genomic breeding values for WSSV resistance.
Two G1 populations were produced, one from G0 breeders with high and the other with low estimated breeding values. A third population was produced from “random” mating of the parent stock. The average survival was 25 percent in the low, 38 percent in the random and 51 percent in the high-genomic breeding value groups. The resulting realized genetic gain and high heritability clearly demonstrate a large potential for further genetic improvement of WSSV resistance in the evaluated L. vannamei population using genomic selection.
Average survival in the G1 population increased from 38 to 51 percent after one generation of genomic selection for high WSSV resistance for the dead or alive (DOA) trait relative to average survival in randomly selected G1 shrimp. Selection for low WSSV resistance gave a very similar response in the opposite direction. This clearly demonstrates the power of genomic selection as a tool to develop WSSV-resistant L. vannamei shrimp using this particular “synthetic” population which contains large variation for WSSV resistance. Due to the large variation in the DOA trait and relatively high heritability, further selection to produce a G2 would be expected to produce greater genetic gains than we evaluated with the production of the G1.
In general, our estimates for heritability of WSSV resistance based on genomics were greater than the estimates obtained from previous conventional evaluations. The higher heritability found in G1 vs. G0 shrimp could be due to a difference in true heritability, because of differing test conditions and/or differing genetic composition between the populations. But it could also be due to estimation inaccuracies with one estimate being closer to the true heritability than the other.
We identified several factors that make genomic selection for WSSV resistance in L. vannamei attractive. First, the methodology based on genomics identified genetic variation in resistance in the initial G0 population. Second, the detection of significant differences between the selection groups shows that selection increased WSSV resistance in this first generation of selection. Third, broad variation was detected within groups. Fourth, the selection response was greater than expected from the selection intensity applied, estimated heritability and other variables. Finally, the 13 percent improved survival in one generation is greater than obtained with conventional selection methodologies, and the genetic gain is commercially interesting.
The survival levels from our results of slightly more than 60 percent in the best populations may not appear as satisfactory for commercial production. However, when vaccination is used as means of protecting populations against epidemics, the population can be effectively protected without vaccinating every individual due to the herd effect. In a similar manner that disease epidemics can be slowed or stopped when a certain level of the population is vaccinated, a certain level of resistant animals in a population, coupled with management practices that are unfavorable to the pathogen and disease, may be sufficient to control WSSV in commercial populations.
We have shown that by using genomic selection we can rapidly increase the level of resistance and hopefully will be able to use this tool to offer growers shrimp populations that can survive and produce in the presence of WSSV without having to totally eradicate the causal organism.
In applying genomic selection for disease resistance, we also need to consider that an unfavorable genetic correlation between growth rate and WSSV resistance exists and that a faster growth rate is desirable for shrimp aquaculture. Simultaneous improvement of both traits could be achieved by combining a fast-growing S-line with high resistance R-line in a multiplier design for commercial broodstock supply. It may also be possible to use genomic selection in such a way that both traits are positively selected, but this needs further investigation.
Another consideration for application is affordability and accessibility. The need for genotyping will add costs to the breeding program, but it may also reduce the need for systematic pedigree recording, which is costly and also induces inefficiencies in the selection process due to separate rearing, as that can be derived from the genotypes. Many genetic markers have already been identified for L. vannamei and technologies for ultra-high throughput genotyping of dense markers are becoming more readily available and less costly to apply.
The other major difficulty faced when selecting for WSSV resistance in shrimp is that it is practically impossible to use survivors of challenge tests as breeding candidates as they can vertically transmit the virus and infect the offspring population. Furthermore, the practical difficulties of cleaning survivors to incorporate them into breeding programs that produce WSSV-specific pathogen-free broodstock are considerable. The practical impossibility of using survivors, and the lack of information on survival performance of breeding candidates and their true genetic relationships with candidates (pedigree relationships have been the standard for previous studies), limits the accuracy of conventional phenotypic selection.
We contend that the ability to evaluate the individual breeding value through genomic selection is the main reason why a higher rate of genetic gain was obtained with genomic selection than that achieved with conventional phenotypic selection of WSSV resistance in L. vannamei. Based on our study results, the realized genetic gain and high heritability clearly demonstrate large potential for further genetic improvement of WSSV resistance in the evaluated L. vannamei population using genomic selection.
Perspectives
Our results show that significant useful genetic improvement for WSSV resistance can be achieved in a breeding program for L. vannamei by applying genomic selection. Compared with conventional methods, the use of genomic data resulted in higher heritability estimates and improved the accuracy of selection to levels that are commercially relevant.
This is the first study with L. vannamei that demonstrates realized genetic gain from genomic selection. Genomic selection is particularly promising for selection programs that challenge live animals with infectious agents when survivors cannot later be used as breeders.
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Author
-
Nicholas Andrew Robinson, Ph.D.
Corresponding author
Breeding and Genetics, Nofima, 1430, Ås, Norway; and
Sustainable Aquaculture Laboratory-Temperate and Tropical (SALTT)
School of BioSciences
The University of Melbourne
Parkville, 3010, Australia
Editor’s note: This article has nine co-authors, but only the corresponding author is listed.[117,97,46,117,100,101,46,98,108,101,109,105,110,117,64,49,110,46,110,111,115,110,105,98,111,114]
Tagged With
Related Posts
Health & Welfare
A look at aquaculture genomics
Advances in genomics assist aquaculture science by deepening the understanding of adaptation, physiology and quantitative genetics.

Innovation & Investment
AquaGen CEO: Genomics are transforming aquaculture
The CEO of AquaGen knew that the Norwegian research group’s work in genomics was key to the salmon industry’s future. And that was before she even worked there.

Health & Welfare
Aquaculture genomics: Progress in identifying species genetics continues
The continued application of genome research to aquaculture will provide unprecedented accuracy for genetic selection of performance and production traits.

Health & Welfare
Barcoding, nucleic acid sequencing are powerful resources for aquaculture
DNA barcoding and nucleic acid sequencing technologies are important tools to build and maintain an identification library of aquacultured and other aquatic species that is accessible online for the scientific, commercial and regulatory communities.


