Significant expansion of gene families was observed in the genome related to locomotion, vision and neural transmission

A genome is all the genetic information of an organism. A high-quality reference genome is crucial for genetic improvement of species and the exploration of the molecular mechanisms underlying important biological processes. However, shrimp genomes contain numerous repetitive sequences, rendering assembly a challenge. Recently developed third-generation sequencing technologies have helped alleviate assembly difficulties caused by heterozygosity and repetitive sequences in genomes.
The genomes of several shrimp species of commercial importance – including black tiger shrimp (Penaeus monodon), Chinese white shrimp (Fenneropenaeus chinensis), Kuruma shrimp (Marsupenaeus japonicus) and others – have recently been assembled using these newer third-generation sequencing technologies. However, compared to other species, the availability of shrimp genomes is still limited.
A draft genome of L. vannamei has been previously generated, although it is highly fragmented and is not constructed at the chromosomal level. The lack of a high-quality genome impedes further genetic research on L. vannamei.
This article – summarized from the original publication (Peng, M. et al. 2023. A high-quality genome assembly of the Pacific white shrimp (Litopenaeus vannamei) provides insights into its evolution and adaptation. Aquaculture Reports Volume 33, December 2023, 101859) – reports on a study to generate a high-quality genome assembly of L. vannamei and provide critical new genomic information for shrimp breeding and investigations of this shrimp species.
Study setup
A de novo assembly refers to the genome assembly of a novel genome from scratch without the aid of reference genomic data. For sequencing and assembly of the genome, L. vannamei samples were obtained from the Shrimp Genetic Breeding Center at the Guangxi Academy of Fisheries Sciences (Nanning, China). Genomic DNA was extracted from muscle tissue and genomic libraries using a commercial kit (SMRTbell kit, PacBio, USA) according to the manufacturer’s protocol and sequenced on the PacBio Sequel platform. This DNA was then processed and analyzed in detail.
For detailed information on the sequencing and assembly of the L. vannamei genome; gene annotation; repeat sequence annotation; transcriptomic sequencing of various tissues; and evolutionary analysis, refer to the original publication.
Results and discussion
L. vannamei has received widespread research attention due to its enormous economic value. A draft genome of this species was previously published, although this earlier genome assembly remains relatively fragmented and incomplete, limiting its applicability as a reference for breeding and research.
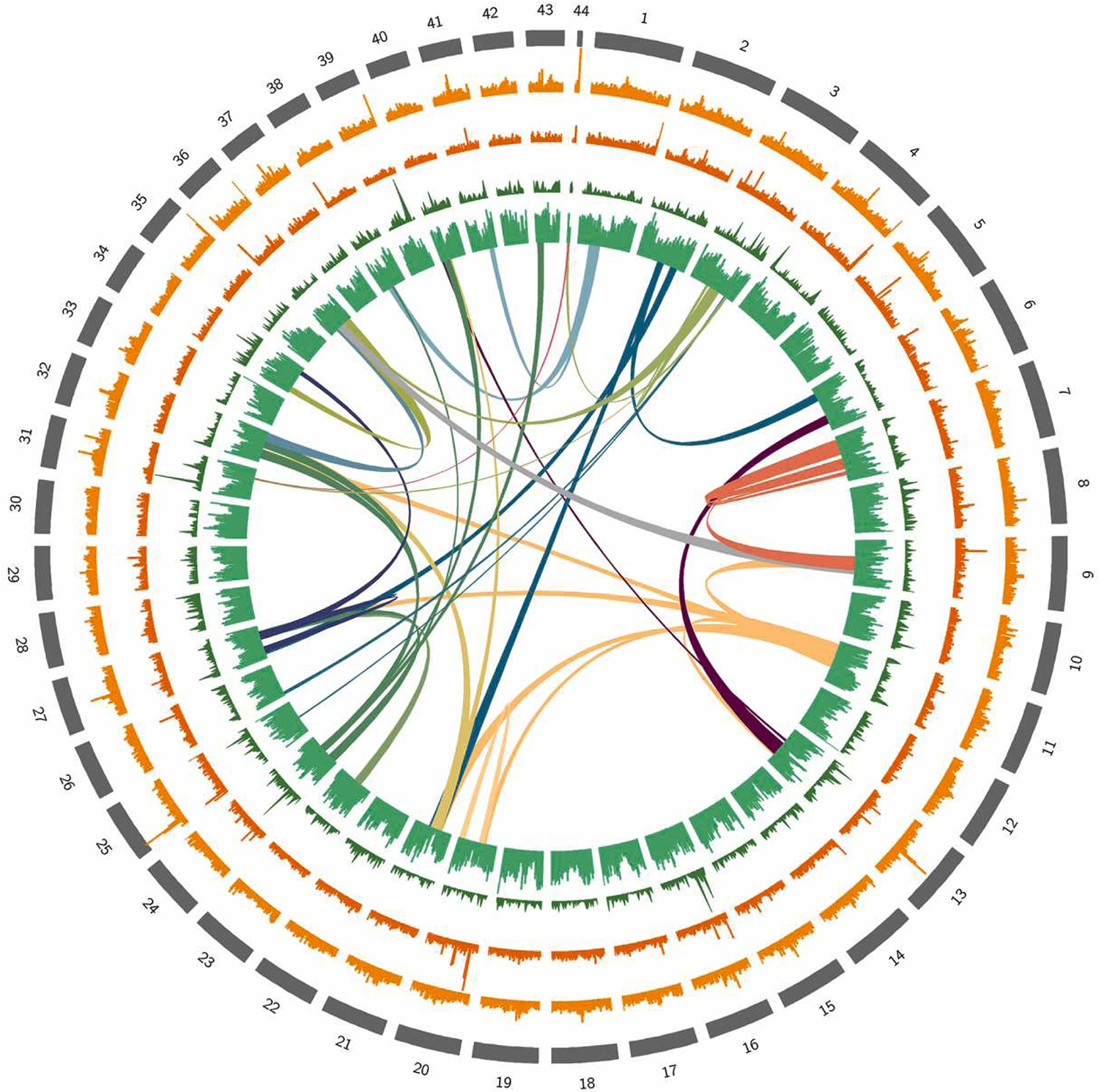
To obtain a resolved genome of L. vannamei, we used novel technologies in this study to generate a high-quality de novo assembly of the genome, with a total length of 1869 megabases (Mbp; megabases or millions of base pairs, is one of the ways used to measure the size of a genome) and other improved characteristics. These metrics considerably improve on the previously published incomplete genome assembly.
This improved genome assembly provides a more comprehensive and accurate representation of the L. vannamei genome, enabling deeper insights into its genetic composition and regulatory mechanisms. It also opens up new opportunities for research and applications. Researchers can now explore the gene content, evolution and functional aspects of L. vannamei in greater detail, including investigations into key traits such as growth, reproduction, disease resistance and environmental adaptation.
Additionally, this high-quality genome assembly serves as an invaluable reference for future studies on L. vannamei genomics, allowing the identification and annotation of previously unknown genes and regulatory elements and facilitating comparative genomics studies to understand evolutionary relationships and the genetic basis of phenotypic diversification within the shrimp family.
While previous research has suggested that the haploid (single set) chromosome count of L. vannamei is 44, more research may be necessary to precisely determine the exact number of chromosomes in this species.
Regarding the annotation of repetitive sequences (short or long patterns of nucleic acids – DNA or RNA – that occur in multiple copies throughout the genome), our results showed a lower proportion compared to a previous study. This variation could stem from the utilization of improved genome assembly techniques in our research or the possibility of missing repeats in our assembly.
With tools like CRISPR, can genome editing deliver more resilience for aquaculture?
Repetitive sequences play a crucial role in genome structure and function, as they are known to regulate gene expression in F. chinensis and L. vannamei, driving adaptive evolution, such as the variable osmoregulatory capacity of these shrimps under low salinity stress. Future studies should focus on addressing the limitations in our assembly to enhance our understanding of the repetitive content within the L. vannamei genome.
Expansion or contraction of gene families are key mechanisms in organismal adaptive evolution. We observed significant expansion of some gene families in the L. vannamei genome. Expanded gene families included some that are primarily involved in movement functions, in addition to other gene families involved in visual and neural transmission functions. The significant expansion of these gene families could lead to enhanced visual, neural speed, and movement capabilities of L. vannamei, perhaps enabling their adaptation to escape predation in dark benthic habitats. The abundant gene repertoire in the L. vannamei genome related to visual, neural transmission and movement may explain their benthic adaptations.
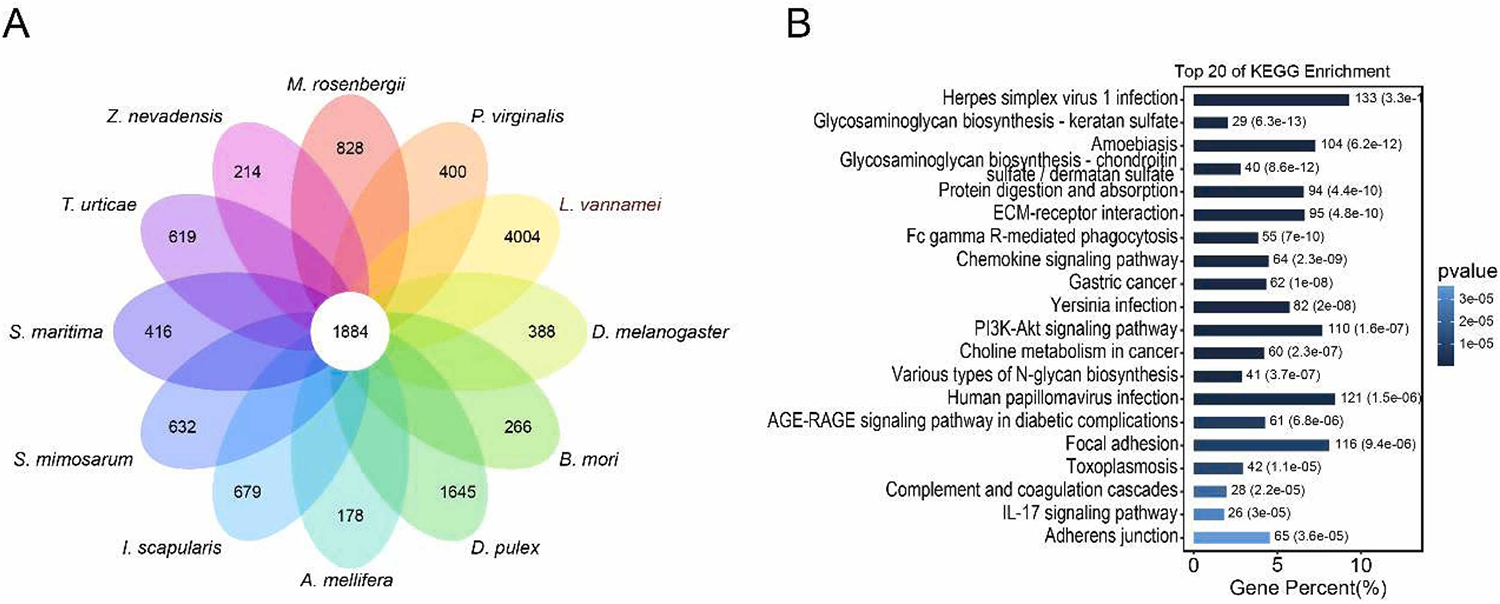
We also constructed a phylogenetic tree involving L. vannamei and 11 other arthropod species. Our findings suggest that L. vannamei is closely related to other crustaceans like the marbled crayfish (Procambarus virginalis) and the giant freshwater prawn (Macrobrachium rosenbergii) and supporting previous studies. However, it is noteworthy that the water flea (Daphnia pulex), another crustacean, did not cluster with L. vannamei, and this may be due to its small genome size of only 200 Mbp, resulting in significant genetic differences compared to L. vannamei with a much larger genome size. This result contradicts the results of previous research and further investigation is required in future studies.
Perspectives
This study documented the generation of a high-quality de novo assembly of the L. vannamei genome, addressing the limitations of previous fragmented assemblies. Our refined product not only provides a more accurate representation of the L. vannamei genome but also offers new avenues for functional genomics, breeding programs and comparative evolutionary studies in this economically significant species.
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Author
-
Dr. Yongzhen Zhao
Corresponding author
Guangxi Key Laboratory of Aquatic Genetic Breeding and Healthy Aquaculture, Guangxi Academy of Fishery Sciences, Nanning 530021, China[109,111,99,46,108,105,97,109,116,111,104,64,111,97,104,122,110,101,104,122,103,110,111,121]
Related Posts

Health & Welfare
Auburn researchers map the blue catfish genome
The work will aid genetic improvement using gene-editing or genome-assisted selection and result in better breeds for the U.S. catfish industry.

Health & Welfare
Australian research partnership maps the black tiger prawn genome
A collaboration of research institutes and industry has mapped the genome of an iconic Australian seafood species, the black tiger prawn.
Health & Welfare
A look at aquaculture genomics
Advances in genomics assist aquaculture science by deepening the understanding of adaptation, physiology and quantitative genetics.

Innovation & Investment
AquaGen CEO: Genomics are transforming aquaculture
The CEO of AquaGen knew that the Norwegian research group’s work in genomics was key to the salmon industry’s future. And that was before she even worked there.



