The Infectious Hypodermal and Hematopoietic Necrosis Virus is established in shrimp ponds in Ecuador

Over the past several years the Infectious Hypodermal and Hematopoietic Necrosis Virus (IHHNV), a shrimp pathogen listed by the World Organisation for Animal Health [WOAH; founded as the Office International des Epizooties, OIE] has been reported as the cause of poor growth in black tiger shrimp (Penaeus monodon) and Pacific white shrimp (Penaeus vannamei) by various publications in Australia, India and, most recently, Ecuador.
The issue we intend to explore here is: “What is the pattern of IHHN virus, IHHNV endogenous viral elements (EVEs, DNA sequences derived from a virus and present in the germline of a non-viral organism) and IHHN PCR test negative shrimp in shrimp ponds in Ecuador, and what do these patterns imply regarding tolerance to this virus in shrimp farms in Ecuador.”
IHHNV, first introduced in the mid-1980s into Ecuador via P. monodon imported from Asia, is well established and indigenous in wild shrimp in the Americas from Peru to the northern reaches of the Gulf of California, as well as being reported from wild shrimp collected from the Gulf of Mexico.
Evolutionarily speaking, Crustacea first appeared around 455 million years ago. The Penaeidae family of shrimp, as members of the Dendrobranchiata (a suborder of decapods, commonly known as prawns), represent examples of eons of successful evolutionary adaptation via generational-mediated mechanisms. Apparently, one such mechanism is the incorporation of DNA sequences that mirror pathogen sequences within the host genome, leading to defense strategies mediated by host RNA.
Naturally occurring shrimp resilience has long been recognized by shrimp farmers and in particular producers of L. vannamei in Latin America, where their capacity to adapt to pathogens is reflected in the use of the management strategy known by the acronym APE for “All Pathogens Exposed.” Additionally, tolerance to selected shrimp pathogens achieved through selective breeding of specific pathogen-free (SPF) shrimp populations in modern shrimp genetics companies is also well established.
Over the past decade or so, various researchers – particularly in Thailand under the direction of Prof. T.W. Flegel and by Dr. V. Alday-Sanz and associates elsewhere – have documented the mechanics of the processes involved at the molecular level as well as demonstrated the utility of selection of SPF shrimp by individual animal testing to establish pathogen-tolerant and pathogen-free L. vannamei derived from Ecuador and transferred to other areas of the world. Their work has been documented extensively and stands as an important foundational contribution to the understanding of how these economically important crustaceans have evolved and thrived.
Applying standard PCR methodology using OIE-listed PCR primers, in this article we report distribution patterns, as measured using standard PCR methods, of embedded IHHNV DNA sequences (EVEs), (probable) actual IHHNV infection and IHHNV PCR test negative shrimp collected from ponds in Ecuador in 2021. We also discuss the relevance of the laboratory findings as these relate to the status of IHHNV diagnosis in farmed shrimp in this region. These results support the conclusion that IHHNV is not a significant pathogen within the gene pools of L. vannamei examined in our study. Additionally, our data indicate that currently applied OIE-listed PCR primers and methodology are out of date because of the presence of EVEs within farmed shrimp.
A recent article on IHHNV in Ecuador briefly highlighted tolerance as demonstrated by minimal IHHN virus-induced tissue pathology in IHHN virus-infected shrimp while simultaneously reporting high titer of IHHNV in the tissue samples. Fundamentally speaking, these two interpretations are not compatible, since IHHN virus replication within cells produces anatomical alterations easily visible in stained histology tissue sections.
Our article’s purpose is to provide a different interpretation based on the presence of IHHNV EVEs as well as provide additional information to bolster that over generational time shrimp and the pathogens that affect them continuously modify their host/pathogen relationships, all part and parcel to the highly evolved defense and self-preservation mechanisms within a population of shrimp.
Study design
Samples of shrimp were collected from 134 (33 nursery and 101 grow-out) ponds in farms along Ecuador’s Pacific Coastline in 2021. Shrimp in the samples from grow-out ponds were weighed to 0.1 grams, dissected and gill/pleopod tissue were removed and preserved in 95 percent ethanol from each animal. Nursery pond samples were not weighed when tissue samples were collected. PCR tests were conducted by staff in the R&D unit of the Ecuadorean shrimp company Empagran S.A.
Each sample was extracted and assayed using the 309F/R, 389F/R, 392F/R 77012/77353, MG831F/R PCR primers according to OIE listed methods. These primers are those listed in the OIE Aquatic Manual for HHNV standard PCR detection of IHHNV. Sequence analyses were not performed on any of the samples in this study.
Results and discussion
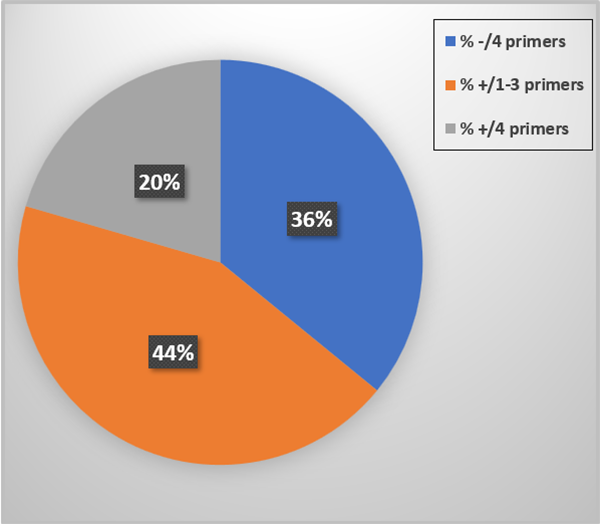
The PCR test outcomes are summarized in Fig. 1. Thirty-six percent of the tissue samples did not prime up with any of the primer sets and 44 percent reacted to one-three of the four primer sets. These represent EVEs, DNA sequences embedded into the host genomic DNA. Finally, 20 percent were PCR test positive for the four primer sets applied. Since EVEs and live IHHNV can occur together in the same shrimp we interpret these animals to be “suspect” PCR test positive for IHHNV (live virus). None of the tissue samples reacted with the MG831F/R primers.
EVEs are foreign DNA sequences embedded in the shrimp host’s DNA. As such, EVEs pass from one generation of shrimp onto their offspring according to the Principles of Mendelian Genetics. As EVEs are within the DNA of every cell in the body of a shrimp, we predict that quantitative PCR testing will yield a low Cq (Quantification Value), an indication that the target DNA sequence is abundant in the sample. If the possibility that an EVE is driving the qPCR reaction is not taken into consideration, then the low Cq value may be misinterpreted to indicate that the signal reflects an abundance of live virus in a sample when the “high titer” could also be just as well explained by the presence of an EVE and/or if a live virus was present, along with an EVE, its presence could be low but appears high because of the contribution of the EVE to the overall sequence reacting in the qPCR protocol.
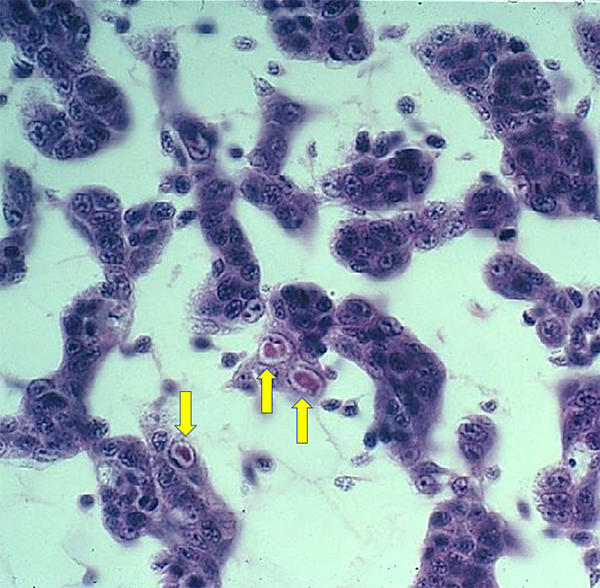
As shown previously, live IHHNV in shrimp tissue samples – when tested using multiple PCR primers – will primer up with all the primers because they all must be present for a functional virus to replicate. Whereas, if when using a panel of primers, if some but not all primers amplify, this pattern is a clear indication that the PCR primers are measuring an EVE and not the complete virus. Furthermore, IHHNV replication results in anatomic alterations within target cell nuclei visible with H&E stain (see Fig. 2) or gene probe methods. A high titer of live virus will be accompanied by a high abundance of anatomically altered host cells, and IHHNV disease symptoms are related to the abundance of infected, anatomically altered host cell loss of normal function.
The distribution of PCR negative, EVE positive (1-3 primers) and IHHNV suspect (4 primers) within groupings organized by shrimp weight within a size range are summarized in Table 1.
Brock, IHHNV, Table 1
| Primer | Primer | Primer | Primer | Average | |
|---|---|---|---|---|---|
| Wt. Group | 309 | 392 | 389 | 77012 | – |
| <1g | 1 | 0.83 | 1 | 0.65 | 0.87 |
| 1-5g | 1 | 0.83 | 1 | 0.44 | 0.82 |
| 5-10g | 0.79 | 1 | 1 | 1 | 0.81 |
| >10g | 1 | 0.78 | 1 | 0.46 | 0.81 |
The above calculations were made by eliminating all the non-PCR primer reactor shrimp to consider only those animals whose tissue sample primed up with one or more of the four primer sets. Within the subset of IHHNV primer reactor shrimp, organized by weight grouping, on average for the four size groups, the prevalence of positive reactors was quite similar (+/- ~6 percent) between the four size groups. The data organized this way does not support a conclusion of an obvious positive relationship between PCR primer positive status vs. shrimp weight grouping.
The OIE Aquatic Manual (Chapter 2.2.4. – Infection with Infectious Hypodermal and Haematopoietic Necrosis Virus, Section 7. Corroborative diagnostic criteria provide guidance for discrimination between a suspect case and a confirmed case) provides benchmarks quoted below:
7.1. Definition of a suspect case: Infection with IHHNV is suspected if at least one of the following criteria is met: i) Clinical signs indicative of infection with IHHNV or ii) Histopathology indicative of infection with IHHNV or iii) Positive result by PCR.
7.2. Definition of a confirmed case: Infection with IHHNV is confirmed if two of the following criteria are met: i) Positive result by in-situ hybridization; ii) Positive result by PCR (always genotype specific); and iii) Sequence analysis to confirm IHHNV nucleic acid sequence.
We agree with others that the current, molecular OIE protocols for IHHNV are out of date from the perspective of EVEs, and that changes are necessary to avoid additional incorrect etiology diagnoses as this relates to IHHNV disease in farmed penaeid shrimp. Although there is available quality, peer-reviewed scientific publications regarding EVEs and proposed tolerance mechanisms at the molecular level, methodologies designed years ago and adopted into the OIE Aquatic Manual for IHHNV to address the EVE issue are inadequate today and lead to misdiagnoses of the current importance of IHHNV to shrimp health.
For example, the OIE states: “In the event that results are ambiguous using the 389F/R “universal” primer set, it is recommended to use primers from a different region of the genome for confirmation testing. In this case that would mean 77013/77353 or 392F/R primer sets for confirmation.” Based on the results of our study, this statement is no longer correct, and we recommend a revision of the protocols.
The primer set MG831F/R was not detected in any of the 134 tissue samples surveyed in this study. Based on these findings the MG831F/R primers were not shown to be useful for IHHNV detection in L. vannamei.
Perspectives
As EVEs are within the host genome and the host genome is within every cell in the body of the shrimp, an EVE or EVEs, if present and measured, would be expected to yield a low Cq value and therefore be interpreted as a tissue sample with high titer of live virus in the host tissue in scenarios where EVEs are not present. However, if EVEs exist then these sources of DNA sequences with the host’s genome must also be taken into consideration as a possible alternate explanation. The PCR data reported herein are a good example of the validity that this is correct.
Therefore, measuring IHHNV associated with shrimp that display slow growth without establishing that the shrimp involved also have a high frequency of IHHNV-infected cells in target organs, or performing sequence analyses to confirm the virus’s genome is present brings with it a level of uncertainty and may result in an incorrect etiologic diagnosis. Although there is confusion with interpretation due to EVEs with current listed OIE IHHNV PCR primers, there is also misinformation because diagnosticians and researchers may not follow the OIE standards as this pertains to a suspect vs. a confirmed case because they fail to carry out OIE confirmation steps to validate their suspect status IHHNV results.
Currently, IHHNV is still an OIE-listed pathogen and the importation of live shrimp is disallowed by entry authorities in most countries. As PCR methods currently listed in the OIE Aquatic Manual are unable to discern between live viruses and EVEs, unless sequencing of IHHNV is carried out, we believe OIE needs to take action to rectify this problem.
In conclusion, should shrimp farmers in Ecuador be concerned about the impact of IHHNV on production on their farms? We do not think so. Moreover, the OIE should de-list IHHNV as a reportable pathogen of penaeid shrimp or, at least revise the PCR methods presently listed for IHHNV in the Aquatic Manual.
Editor’s Note: This article was updated to include the fact that IHHNV was first introduced in the mid-1980s into Ecuador via P. monodon shrimp imported from Asia, not L. vannamei shrimp. We regret the error.
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Authors
-
James A. Brock, D.V.M, retd.
Corresponding author
95-430 Kamahana Place, Mililani, Hawaii USA[109,111,99,46,114,114,46,105,105,97,119,97,104,64,49,48,48,106,107,99,111,114,98]
-
Pablo Intriago, Ph.D.
South Florida Farming Corp., Southwest Ranches, Florida USA; and Empagran, ABA Division Balanceado, Guayaquil, Ecuador
-
Andres Medina, M.Sc.
Empagran S.A. Semacua Division Laboratories, Guayaquil, Ecuador
Tagged With
Related Posts

Health & Welfare
Current status of IHHNV infection in Peru’s and Ecuador’s shrimp industries
The virus-host relationship, where circulating genotypes of IHHNV co-exist with farmed Pacific white shrimp, is potentially evolving, authors find.

Health & Welfare
A study of Zoea-2 Syndrome in hatcheries in India, part 1
Indian shrimp hatcheries have experienced larval mortality in the zoea-2 stage, with molt deterioration and resulting in heavy mortality. Authors investigated the problem holistically.

Aquafeeds
Biosecurity protocols needed for shrimp feeds, feeding practices
Shrimp aquafeeds – live, fresh or formulated – should not be an entry point of potential pathogens to the shrimp and/or to their culture systems.
Health & Welfare
Emerging disease: Shrimp Hemocyte Iridescent Virus (SHIV)
SHIV is a new Pacific white shrimp virus in the Iridoviridae family. Authors also developed an ISH assay and a nested PCR method for its specific detection.



