Polymerase chain reaction confirms presence

Shrimp culture is one of the major income sources in the northwest region of Mexico. In the state of Sinaloa, 7,000 people from 10 different districts along the coast are employed at 430 shrimp farms that cover approximately 42,000 hectares.
A major problem for the shrimp industry has been the disease caused by the white spot syndrome virus (WSSV), which affects a broad range of hosts and has the ability to spread among different species. Due to the prevalence of the virus and the high mortality rates it causes, Sinaloa’s shrimp production has decreased significantly during the last decade.
Because of the impacts of WSSV on shrimp farming in Mexico, a joint effort involving the government, the shrimp industry and the academic community are working to develop strategies to attenuate the effects of the disease by implementing biomonitoring programs and biosecurity measures to control or eliminate the dispersion of the virus.
WSSV variants
The causal agent of the disease is a dsDNA virus whose genome has been fully sequenced. Although two different WSSV profile types have been detected, it was not known whether different variants were present in the farms and estuaries in Sinaloa.
In order to answer this question, the authors recently carried out a study using the ORF94 method to identify variants of WSSV in the region. The work was supported by grants from Fundación Produce Sinaloa and the main author.
Study setup
A total of 133 WSSV-positive shrimp specimens from 11 shrimp farms in Sinaloa were analyzed. For DNA extraction, 25 mg of both fresh and 70 percent ethanol-preserved gill samples were used. DNA was isolated using a commercial tissue kit.
The presence of WSSV was confirmed by polymerase chain reaction (PCR). To amplify the ORF94 gene, the methodology reported by Chainarong Wongteerasupaya in 2003 was applied. The identity of PCR products was confirmed by DNA sequencing using a commercial sequencer and sequencing kit with a commercial sequencing software program.
Results
WSSV-positive specimens were clearly identified by PCR. Eight genotypes were recognized (Table 1). The most common genotype had nine tandem repeats (PCR fragment of 670 bp), followed by the one with seven tandem repeats (562 bp). Both genotypes sed a high prevalence, even 100 percent in some sites (Figs. 1 and 2).
Morales, Number of tandem repeats and corresponding PCR products, Table 1
| Tandem Repeats | PCR Product (bp) |
|---|---|
| 6 | 508 |
| 7 | 562 |
| 8 | 616 |
| 9 | 670 |
| 10 | 724 |
| 11 | 778 |
| 12 | 832 |
| 13 | 886 |
| 14 | 940 |
| 15 | 994 |
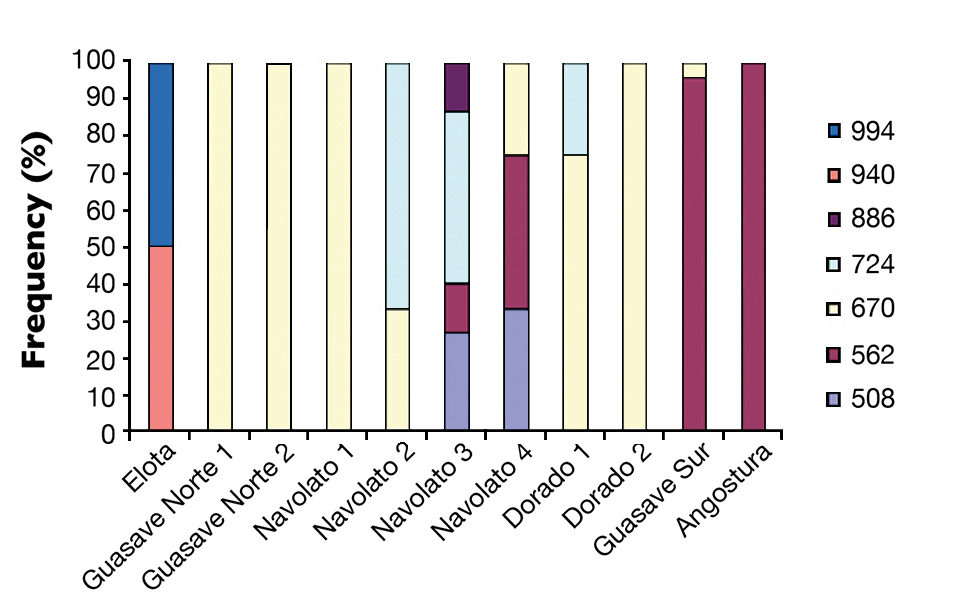
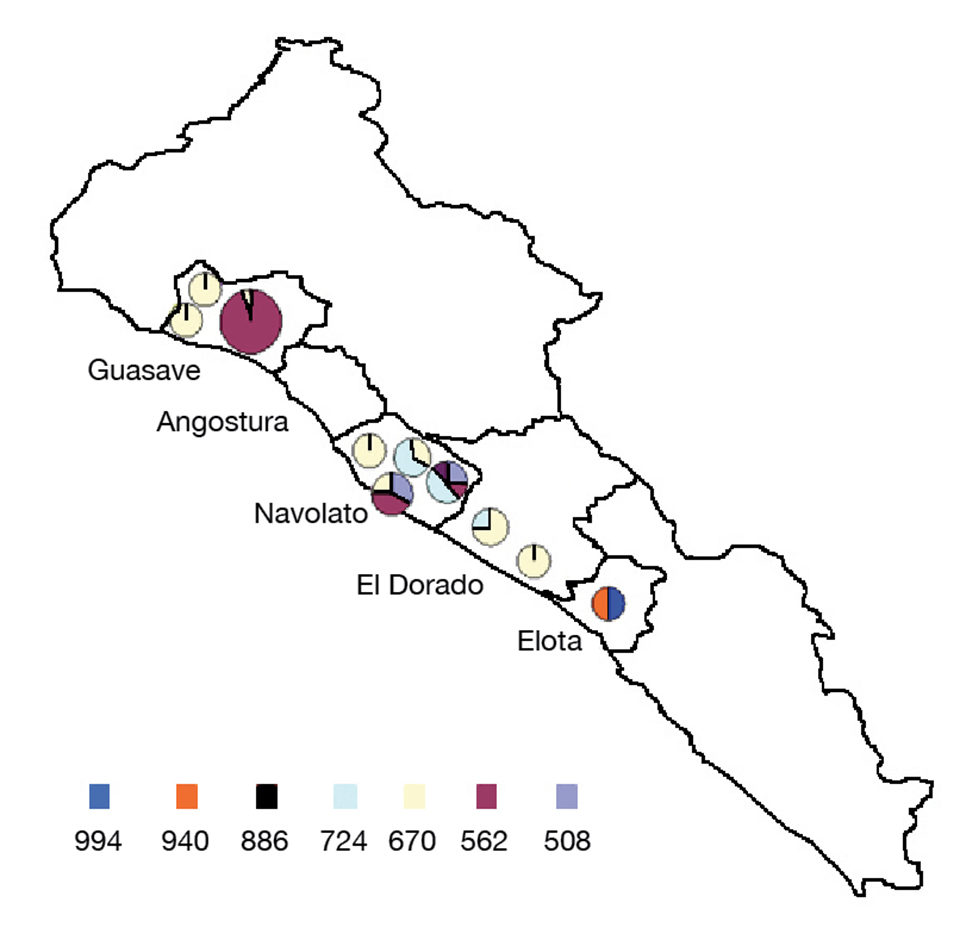
The genotypes with 14 and 15 tandem repeats (940 and 994 bp, respectively) were found only in the district of Elota with a prevalence of 50 percent each. Genotypes with 6, 10 and 13 tandem repeats were found in Navolato with a lower prevalence, and one genotype with 10 tandem repeats (724 bp) was found in El Dorado with a prevalence of 25 percent. Higher variability of genotypes was observed in Navolato 3 and 4, whereas several districts presented only one genotype.
The ORF94 gene has already been used to genotype WSSV in shrimp. For instance, in genotyping shrimp from Thailand, Wongteerasupaya found 12 genotypes with six to 20 tandem repeats in 2000 to 2002. The most common genotype had eight tandem repeats, followed by those with six and nine tandem repeats. Genotypes present in low frequencies had 10, 14, 19 and 20 tandem repeats.
In the authors’ study, PCR products containing more than 15 tandem repeats were not found. Interestingly, in some cases, a faint band of different size was observed together with a more intense band. According to Wongteerasupaya and co-workers, this could be the result of simultaneous infection by two WSSV variants.
Variant distribution
Study results indicated that the distribution of WSSV variants related to their locations. Variants with seven tandem repeats were found on the northern side of the state, and variants with 14 and 15 tandem repeats appeared on the southern side. Variants with nine tandem repeats were common at almost all of the 11 sampled sites.
(Editor’s Note: This article was originally published in the November/December 2009 print edition of the Global Aquaculture Advocate.)
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Authors
-
Dr. María Soledad Morales-Covarrubias
Centro de Investigación en Alimentación y Desarrollo A.C.
Unidad Mazatlán en Acuicultura y Manejo Ambiental
Sábalo Cerritos s/n A.P. 711
C.P. 82010
Mazatlán, Sinaloa, México[120,109,46,100,97,105,99,64,108,111,115,105,114,97,109]
-
Rubí Hernández-Cornejo
Centro de Investigación en Alimentación y Desarrollo A.C.
Unidad Mazatlán en Acuicultura y Manejo Ambiental
Sábalo Cerritos s/n A.P. 711
C.P. 82010
Mazatlán, Sinaloa, México -
Alejandra García-Gasca
Centro de Investigación en Alimentación y Desarrollo A.C.
Unidad Mazatlán en Acuicultura y Manejo Ambiental
Sábalo Cerritos s/n A.P. 711
C.P. 82010
Mazatlán, Sinaloa, México
Tagged With
Related Posts

Responsibility
A look at various intensive shrimp farming systems in Asia
The impact of diseases led some Asian shrimp farming countries to develop biofloc and recirculation aquaculture system (RAS) production technologies. Treating incoming water for culture operations and wastewater treatment are biosecurity measures for disease prevention and control.

Health & Welfare
A study of Zoea-2 Syndrome in hatcheries in India, part 1
Indian shrimp hatcheries have experienced larval mortality in the zoea-2 stage, with molt deterioration and resulting in heavy mortality. Authors investigated the problem holistically.
Health & Welfare
Alphavirus replicon particles potential method for WSSV vaccination of white shrimp
A study demonstrated that VP19 and VP28 white spot syndrome virus envelope proteins expressed by replicon particles provided protection against mortality due to WSSV in shrimp.

Health & Welfare
Asepsis key to prevent contamination in shrimp hatcheries
Maintaining biosecurity and asepsis in larval shrimp production is a key component of the production chain in Ecuador, which requires the production of 5.5 billion larvae monthly from 300-plus hatcheries.


