For routine diagnostics and low-grade infection detection

The microsporidium Enterocytozoon hepatopenaei (EHP) infects penaeid shrimp and has been considered a critical threat to shrimp aquaculture, causing serious economic losses at shrimp farms in several Asian countries. The target organs include the hepatopancreas and midgut, so EHP infection can affect digestive and absorptive function, resulting in growth retardation. Shrimp could be infected with EHP by cannibalism or by feeding on EHP contaminated live feeds.
EHP diagnosis methods targeting SSU rRNA sequence
Several EHP diagnostic methods have been developed based on the small subunit (SSU) rRNA, including PCR, qPCR, in situ hybridization, and loop-mediated isothermal amplification assays. However, SSU rRNA sequences are not highly specific in discriminating microsporidia and SSU rRNA-based PCR methods may generate false positives in non-shrimp samples. For example, the EHP’s SSU rRNA sequence shared a 90 percent similarity to that of Enterospora canceri that infects marine crabs. Therefore, a more specific PCR method is needed for EHP diagnostics.
New EHP diagnosis method targeting EHP β-tubulin gene
Consequently, we developed a new EHP diagnosis method with two sets of primers designed based on the β-tubulin gene sequence (GenBank No. KX842357, Table 1).
Han, EHP detection, Table 1
| Primer name | Sequence (5’ to 3’) | Amplicon size (bp) |
|---|---|---|
| β-tubulin 1st-step PCR | ||
| EHP-618F | CAGCTGGTTGAAAATGCAAA | 618 |
| EHP-618R | GTGCAAAAATGCCTTTCGTT | 618 |
| β-tubulin 2nd-step (nested) PCR | ||
| EHP-237F | GATATGCGCCTCTGTGTTCA | 237 |
| EHP-237R | TGTTTGGAATCCACTCGACA | 237 |
A first-step PCR was carried out with the primers EHP-618F/R under the following cycling conditions: initial denaturation at 94 degrees-C for 3 minutes, followed by 35 cycles of 94 degrees-C for 30 seconds, 58 degrees-C for 30 seconds, and 72 degrees-C for 30 seconds, and a final extension at 72 degrees-C for 7 minutes.
Then, a second-step (nested) PCR was carried out with the internal primers EHP-237F/R under the following cycling conditions: initial denaturation at 94 degrees-C for 3 minutes, followed by 20 cycles of 94 degrees-C for 30 seconds, 58 degrees-C for 30 seconds, and 72 degrees-C for 30 seconds, and a final extension at 72 degrees-C for 7 minutes.
The second step (nested) PCR is 100-fold more sensitive than the first-step PCR (Fig. 1), and is thus suitable for detecting a low level EHP infection.

Field samples study – first-step PCR assay
To compare the sensitivity in field samples, we applied several EHP-infected shrimp samples (n = 22) collected from Southeast Asian countries (Vietnam, India, Thailand, and Indonesia) and EHP-positive brine shrimp artemia spp. (n = 6; all at adult stage) determined by an EHP SSU rRNA PCR (PCR products correspond to accession no. KP759285) to the first-step PCR assay. After the 1st-step PCR, 618-bp amplicons were generated from all EHP samples (n = 22) from various shrimp farms in the countries named above (Fig. 2).
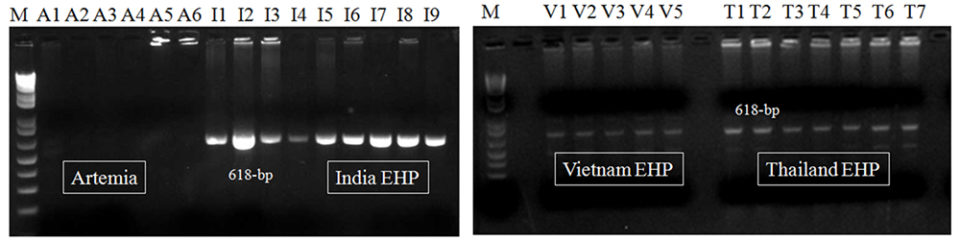
The first-step PCR assay is simple and fast. However, it did not generate any amplicons from the EHP-positive artemia biomass samples (n = 6), which might be due to low quantities of EHP present in these samples (Fig. 2).
Field samples study – second-step (nested) PCR assay
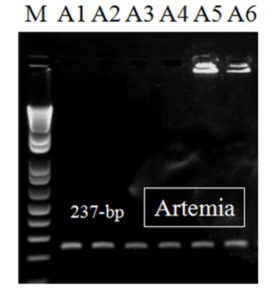
After the second-step (nested) PCR, a DNA fragment of 237-bp was observed in all EHP samples (n = 22), including those from the artemia biomass samples (Fig. 3). Diagnostic results obtained by this nested PCR method are beneficial for monitoring of shrimp with low levels of EHP infection. Also, this method is helpful to detect EHP in live feed (such as artemia), which is important in the field because live feed is a potential risk for EHP infection.
Perspectives
In this study, we developed a nested PCR method using β-tubulin gene sequences. This new PCR method is specific to EHP and can be used in routine diagnostics. Also, this method is useful to detect low-grade EHP infection. Additionally, we detected EHP in artemia samples by this nested PCR method. It suggests the possibility of EHP transfer from the live feed to shrimp broodstock. In fields, shrimp could be infected with EHP by feeding on EHP contaminated live feed such as artemia. Further laboratory experiments are required to figure out the infectivity of the EHP infected artemia in shrimp.
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Authors
-

Jee Eun Han, DVM, Ph.D.
Laboratory of Aquatic Biomedicine
College of Veterinary Medicine
Kyungpook National University
Daegu 41566, Korea[109,111,99,46,108,105,97,109,103,64,51,50,50,49,101,106,110,97,104]
-

Kathy F.J. Tang, Ph.D.
Yellow Sea Fisheries Research Institute
Chinese Academy of Fishery Sciences
Qingdao 266071, China -

Ji Hyung Kim, Ph.D.
Infectious Disease Research Center
Korea Research Institute of Bioscience and Biotechnology
Daejeon 34141, Republic of Korea
Tagged With
Related Posts

Health & Welfare
EHP a risk factor for other shrimp diseases
Laboratory challenges and a case-control study were used to determine the effects of EHP infection on two Vibrio diseases: acute hepatopancreatic necrosis disease (AHPND) and septic hepatopancreatic necrosis (SHPN).

Intelligence
At Aquaculture Roundtable Series, talk of change for Thai shrimp
The theme of the Aquaculture Roundtable Series in Chiang Mai was “Need For Change.” That means innovations in all production phases of Thai shrimp.
Health & Welfare
Microsporidian impacts shrimp production
Enterocytozoon hepatopenaei (EHP), a microsporidian parasite widely found in Asia and other areas, is impacting aquaculture by severely retarding the growth of cultured shrimp. EHP infects the tubules of the hepatopancreas in shrimp, which damages the organ’s ability to gain nutrition from feed.

Health & Welfare
New management tools for EHP in penaeid shrimp
Authors examined the histological features from shrimp infected with the emerging microsporidian parasite Enterocytozoon hepatopenaei (EHP). A PCR assay method was used to detected in hepatopancreatic tissue, feces and water sampled from infected shrimp tanks, and in some samples of Artemia biomass.

