The novel IMNV genotype is more closely related to IMNV strains from Indonesia

The Infectious Myonecrosis (IMN) disease caused by the Infectious Myonecrosis Virus (IMNV) is among many viral diseases of farmed shrimp that have spread around the world and caused significant economic losses. The disease was initially detected in northeast Brazil in 2002 and was later reported from Java Island in Indonesia around 2006.
More recently, IMN has been reported in West Bengal, India. Four species of shrimp have been found susceptible to IMNV, including the Pacific white shrimp (Penaeus vannamei), the Pacific blue shrimp (P. stylirostris), the black tiger shrimp (P. monodon) and the southern brown shrimp (Farfantepenaeus subtilis).
The disease caused by IMNV progresses slowly, with persisting low mortality rates throughout the production cycle and ends with a cumulative mortality of 40–60 percent. Due to its significant negative effect on the farmed shrimp industry and its economic impact, IMNV has been listed as a reportable agent by the World Organization for Animal Health (https://www.woah.org; founded as the Office International des Epizooties, OIE, in 1924).
The economic impact of IMNV in Brazil has been significant and losses have been estimated at up to U.S. $440 million between 2002 and 2005. An urgent update on a more recent production loss figure at a global scale is needed to fully understand and conceptualize the broader impact of IMNV on shrimp aquaculture. Since 2016, unusual mortalities that progress more rapidly and result in higher cumulative mortality of up to 80 percent have been recorded in the States of Para, Maranhão, Piaui, Ceará, Rio Grande do Norte, Alagoas, Sergipe and Bahia in Brazil. Disease diagnostics by molecular and histopathological techniques confirmed the presence of IMNV with a high viral titer in the infected animals. We hypothesized that a new strain of IMNV had emerged in these samples that was possibly more pathogenic causing higher mortalities compared to the past.
This article – adapted and summarized from the original publication [Andrade, T.P.D. et al. 2022. Novel infectious myonecrosis virus (IMNV) variant is associated with recent disease outbreaks in Penaeus vannamei shrimp in Brazil. Aquaculture 554 (2022) 738159] – reports the full-length genome sequence of the novel IMNV strain we determined and report its genetic relation to other IMNV strains from Brazil and Indonesia.
Study setup
In this study, we used Next Generation Sequencing to determine the complete genome of a novel IMNV strain (Br-1) from the State of Ceará from the 2018 culture cycle and associated with unusually high mortalities.
The IMNV-infected P. vannamei examined in this study were collected from 2016 to 2019 from commercial shrimp-producing facilities presenting cumulative mortalities reaching up to 8 percent. These facilities are in the Brazilian Northeast region, where most of the production takes place.
The average weight of juvenile shrimp from affected ponds was 3.0 grams. The animals were collected for molecular biology (pleopods) and for histopathology (abdominal segments displaying muscle necrosis) analyses at the Laboratório de Diagnóstico de Enfermidades de Crustáceos of the Universidade Estadual do Maranhão.
For detailed information on shrimp sampling; detection of IMNV; histopathology; next generation sequencing, and bioinformatic and phylogenetic analyses, refer to the original publication.
Current status of IHHNV infection in Peru’s and Ecuador’s shrimp industries
Results and discussion
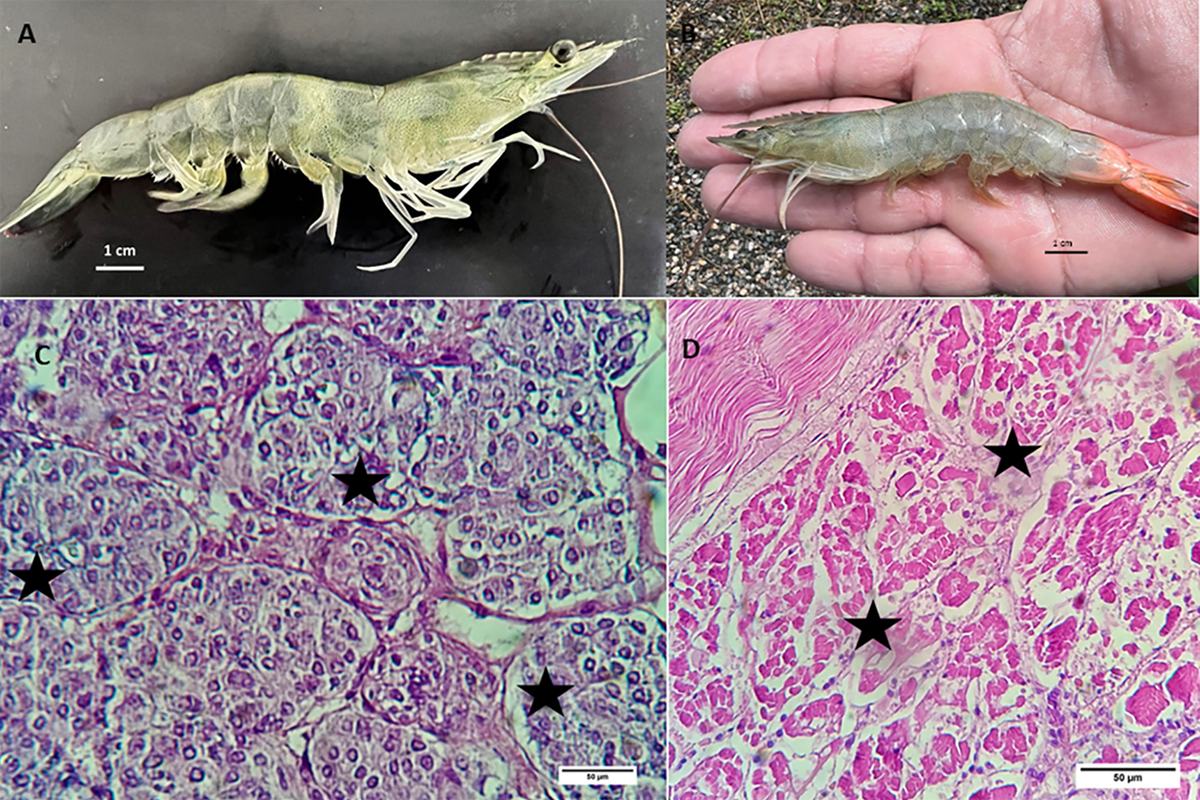
Regarding the screening of P. vannamei samples for infectious IMN and White Spot Diseases, from 2016 to 2019, disease outbreaks were reported from many P. vannamei culture areas in Brazil. In recent years, the country’s shrimp farming industry has suffered major losses due to White Spot Disease outbreaks. Therefore, we initially speculated that the disease outbreaks were due to White Spot Disease (WSD) caused by White Spot Syndrome Virus (WSSV), although the clinical signs in the moribund animals appeared to be more similar to IMN (Fig. 1A). The moribund shrimp displayed signs of muscle necrosis in the distal abdominal segment and uropods (posterior appendages used in locomotion) and reddening of the tail fan (Fig. 1B). These clinical signs are characteristics of IMN. Histologically, 54 four shrimp showed coagulative necrosis of the skeletal muscle accompanied by haemocyte infiltration and fibrosis (Fig. 1D). In addition, lymphoid organ spheroids were also detected in shrimp showing the histological hallmarks of IMNV infection (Fig. 1C).
The RT-PCR results showed the samples had varying levels of infection with IMNV (copy number varies from 26 to 3.6 × 1011) and WSSV was either not detected or detected at a much lower level than IMNV. Following the initial detection of IMNV and WSSV, a subset of samples from the States of Ceará, Maranhão and Piaui were sent to the Aquaculture Pathology Laboratory at the University of Arizona. These samples were again tested for IMNV using a real-time RT-PCR following an OIE-recommended protocol. All samples tested positive for IMNV and only one sample was negative for IMNV, from the State of Ceará.
A subset of samples tested positive for IMNV were sequenced using next-generation sequencing. From our analysis of whole genome sequencing, the identity (percent) between the novel IMNV genome and the reference genome is 97.94 percent, indicating this is a novel strain of IMNV. This novel IMNV strain was designated Br-1, and its whole genome phylogeny showed that it clustered with IMNV strains from Indonesia. The Br-1 full-length genome sequence was deposited in GenBank. This database is an open-access, annotated collection of all publicly available nucleotide sequences and their protein translations. It is produced and maintained by the National Center for Biotechnology Information (NCBI), part of the National Institutes of Health (NIH) in the United States as part of the International Nucleotide Sequence Database Collaboration (INSDC).
The increased risk of transboundary spread of crustacean diseases through the movement of live broodstock and postlarvae has led to the inclusion of many pathogens within international legislative frameworks of the WOAH as diseases of global concern. As a result, crustaceans need to be screened for these diseases before transboundary movement. This is a vital tool for reducing the impact of these pathogens on global production from aquaculture and fisheries.
Since the mid-1980s, the periodic emergence of viral pathogens impacting the global farmed shrimp industry – such as Baculovirus penaei (BP), Infectious Hypodermal and Hematopoietic Necrosis Virus (IHHNV), Taura Syndrome Virus (TSV), Yellow Head Virus (YHV), White Spot Syndrome Virus (WSSV) and more recently IMNV – have seriously affected the industry on a global scale. These viral epizootics have been attributed to the transboundary movement of diseased broodstock, larvae and commodity shrimp.
Brazil study: Beta-glucans improve survival of IMNV-infected white shrimp
Prior to the emergence and subsequent spread of IMNV, many viral shrimp pathogens spread around the world. The emergence of IMNV in Brazil in 2002 and its subsequent spread to Indonesia in 2006 exemplify the “lessons not learned” from previous experiences with the trading of infected broodstock animals.
New-found genotypes (complete set of genetic material) of IMNV continue to emerge as the farmed shrimp industry keeps expanding globally. Recently, three unique IMNV genotypes were reported from the East Java region in Indonesia; these novel IMNV genotypes from Indonesia appeared to be less pathogenic, causing less than 30 percent cumulative mortality in comparison to the 40 to 60 percent mortality expected with IMNV infection during a production cycle. This decreased mortality rate could be associated with mutations in the pathogen genome.
Anecdotal data suggests that over the past few years, there has been the unofficial introduction of some genetic lines such as “Line X” from Indonesia to Brazil. Apparently, these genetic lines have been performing well in IMNV-endemic areas in Indonesia. In addition, “Line X” shows high survival in WSSV-endemic areas in Brazil. When mortalities started occurring in shrimp farms raising some of these lines and located in the States of Maranhão, Piaui and Ceará, there was an urgent need to screen these animals for IMNV and WSSV. The results reported here unequivocally show the animals were acutely infected with IMNV.
Our next-generation sequencing of a representative sample revealed the presence of a novel IMNV strain, Br-1 (IMNV/Ce-2018) that is associated with a higher mortality than previously reported for IMNV strains from Brazil. Interestingly, the phylogenetic analysis (the study of the evolutionary history and relationships among or within groups of organisms) using the full-length genome sequence showed that Br-1 clusters (groups) with IMNV strains from Indonesia. Furthermore, sequence similarity analysis show that strain Br-1 is more similar to Indonesian strains (particularly to strain EF061744.1).
IMNV initially spread from Brazil to Indonesia, and anecdotal evidence suggests that in recent years there has been movement of live animals between the two countries. It remains to be determined if the movement of broodstock between the two countries is contributing to the emergence of novel strains as the animals are exposed to unique culture conditions in two different geographical locations.
Perspectives
In fish and crustacean aquaculture, relocation and transportation of live animals between different areas is a common practice. This practice presents a risk due to the possible dispersion of symbionts, parasites and diseases to unique geographical conditions and exposing naïve populations to exotic diseases or unique strains of existing pathogens in the introduced region.
Based on our study results, we report a novel IMNV genotype from Brazil that is more closely related to IMNV strains from Indonesia based on full-length genome phylogeny. It is tempting to speculate that the re-introduction of an IMNV strain from Indonesia to Brazil might be contributing to the emergence of more virulent strains. However, large-scale sequencing of contemporary IMNV isolates originating from the two countries needs to be conducted to delineate and confirm the origin of these unique strains. Nevertheless, these findings underscore the importance of disease screening of broodstock and postlarvae to prevent the spread of potentially lethal shrimp diseases worldwide.
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Authors
-
Thales P.D. Andrade, Ph.D.
Laboratório de Biopatologia de Organismos Aquáticos, Centro de Ciências Agrárias, Universidade Estadual do Maranhão, Maranhão, Brazil
-
Roberto Cruz-Flores, Ph.D.
Aquaculture Pathology Laboratory, School of Animal & Comparative Biomedical Sciences, The University of Arizona, Tucson, AZ, USA; and Centro de Investigación Científica y de Educación Superior de Ensenada, (CICESE), Ensenada, Baja California, Mexico
-
Hung N. Mai, Ph.D.
Aquaculture Pathology Laboratory, School of Animal & Comparative Biomedical Sciences, The University of Arizona, Tucson, AZ USA
-
Arun K. Dhar, Ph.D.
Corresponding author
Aquaculture Pathology Laboratory, School of Animal & Comparative Biomedical Sciences, The University of Arizona, Tucson, AZ USA[117,100,101,46,97,110,111,122,105,114,97,64,114,97,104,100,97]
Tagged With
Related Posts

Health & Welfare
Hybrid assay can detect IMNV in resource-poor settings
The method can shorten analysis time and has applications for IMNV diagnosis in resource-poor settings because it does not require specialized equipment.
Health & Welfare
Brazil shrimp farm performs genetic selection for IMNV resistance, growth
The Queiroz Galvão Alimentos shrimp farm and hatchery in Brazil have been working with Concepto Azul to implement a disease-prevention and genetic-breeding program that addresses ongoing impacts from infectious myonecrosis virus (IMNV) and other pathogens.

Health & Welfare
A comprehensive look at the Proficiency Test for farmed shrimp
The University of Arizona Aquaculture Pathology Laboratory has carried out the Proficiency Test (PT) since 2005, with 300-plus diagnostic laboratories participating while improving their capabilities in the diagnosis of several shrimp pathogens.

Health & Welfare
Influence of stressors on shrimp susceptibility to White Spot Disease, Part 1
Study stresses the importance of understanding the impact of environmental stressors on farmed shrimp contracting White Spot Disease.



