Frequently monitor wild broodstock to develop SPF shrimp
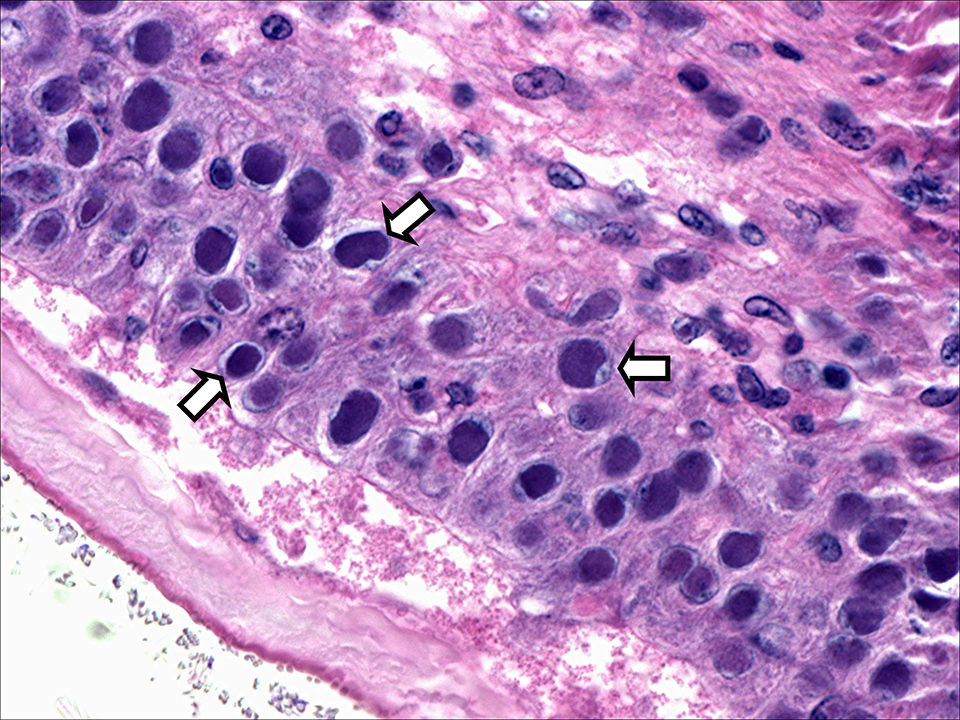
Viral pathogens have caused significant economic losses for the shrimp-farming industry around the world. Among these, white spot syndrome virus (WSSV) and Taura syndrome virus (TSV) are both extremely virulent, and infection can result in high mortalities in populations of farmed shrimp. During 2010 and 2011, both WSSV and TSV were detected in the Kingdom of Saudi Arabia, where they have severely affected production of Indian white shrimp (Penaeus indicus).
To better understand the origins and spread of these viruses within Saudi Arabia, the authors applied molecular genotyping to characterize viral isolates collected from infected populations. WSSV genotyping is based on the variable number of tandem repeat (VNTR) analysis within ORF125 (69-bp per repeat unit) and ORF94 (54-bp per repeat unit). For TSV genotying, the sequence of capsid protein 2 (CP2) coding region that varies among isolates has been used for phylogenetic analysis.
In their study, the authors applied the VNTR analysis to characterize WSSV genotypes among 15 Saudi Arabian isolates and conducted a phylogenetic analysis to determine if eight TSV isolates clustered with the previously determined groups.
WSSV genotyping
Polymerase chain reaction (PCR) testing was performed with primers ORF125-flank F/R, and all 15 samples were detected with specific amplicons at sizes of 652, 722 or 792bp. The results showed three types of repeat units: 6, 7 and 8 (Table 1). VNTR analysis with primers ORF94-F/R also found three types of repeat units: 7, 13 and deletion.
Tang, Number of repeat units, Table 1
| Isolate | Repeat Units in ORF125 | Repeat Units in ORF94 |
|---|---|---|
| SA/10 | 6 | 7 |
| SA/11-1 | 8 | 13 |
| SA/11-2 | 8 | 13 |
| SA/11-3 | 8 | 13 |
| SA/11-4 | 8 | 13 |
| SA/11-5 | 8 | 13 |
| SA/11-6 | 7 | Deletion |
| SA/11-7 | 7 | Deletion |
| SA/11-8 | 7 | Deletion |
| SA/11-9 | 7 | Deletion |
| SA/11-10 | 7 | Deletion |
| SA/11-11 | 7 | Deletion |
| SA/11-12 | 7 | Deletion |
| SA/11-13 | 7 | Deletion |
| SA/11-14 | 7 | Deletion |
Based on these results, the authors tentatively designed the WSSV genotype with the numbers of repeat units in both ORF125-ORF94 as {6125, 794}, {7125, del94} and (8125, 1394}. The subscript numbers indicate the number of repeats in a specific ORF, while “del” stands for deletion.
TSV analysis
For TSV genotyping, eight isolates were extracted for RNA, followed by RT-PCR amplification within the CP2 region. Sequencing of the amplified products found the eight TSV sequences were very close, with a mean distance of 0.7 percent. The Saudi Arabian TSV had a 90.0 percent identity with the reference Hawaii isolate collected in 1994.
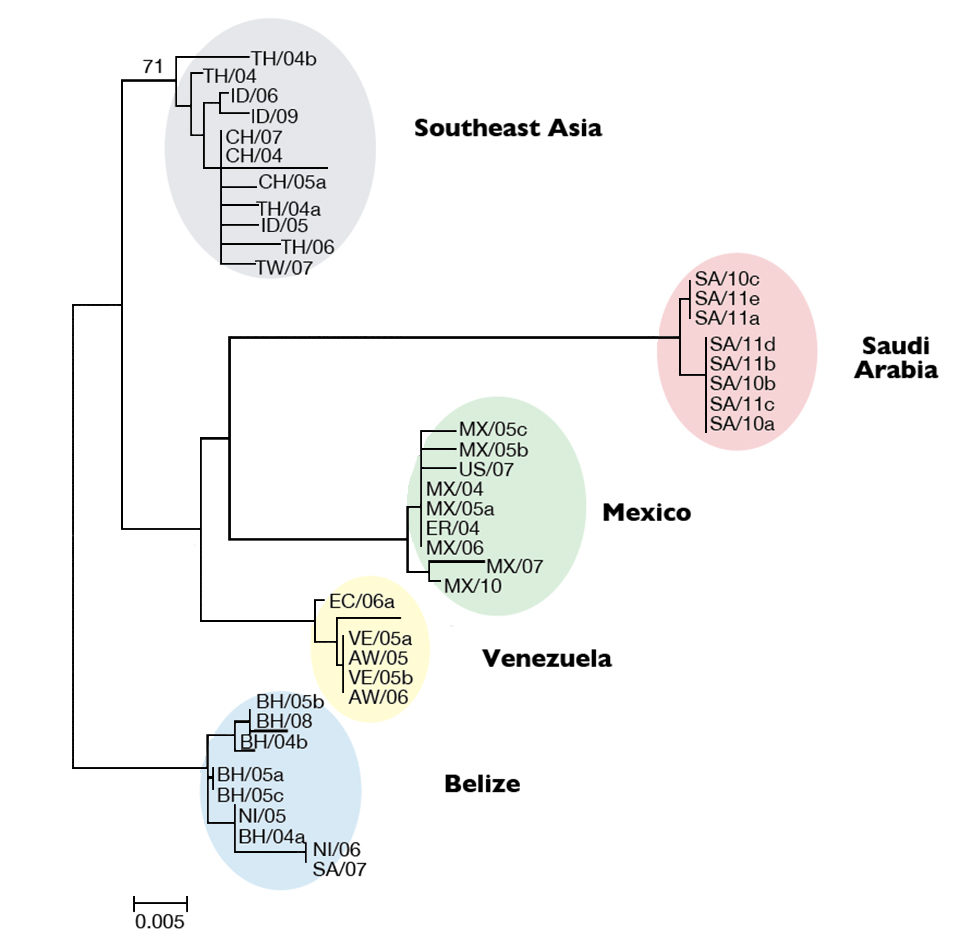
The authors determined its genotype by constructing a phylogenetic tree that compared the Saudi Arabian isolates to 36 isolates from other areas. The result revealed five major TSV lineages (Fig. 1). All eight Saudi Arabian isolates clustered into a distinct group separated from the existing groups from Belize, Southeast Asia, Venezuela and Mexico. The Saudi Arabian lineage was highly supported, and these isolates appeared to have the same origin.
Perspectives
The WSSV genotyping data did not prove particularly useful in determining the origin of WSSV in Saudi Arabia farms. Among six isolates with genotypes of {6125, 794} and {8125, 1394}, their numbers at each ORF were well within the variability reported for isolates from Asia and Latin America. The other nine WSSV isolates were of the newly discovered genotype: {7125, del94}, suggesting they originated from a common, but unidentified, source.
In some outbreaks, the source of virus can be determined by genotyping analysis coupled with the history of shrimp introductions, either live or from the commodity market. The Saudi Arabian genotype with the deletion in ORF94 is unlike those from other geographic areas. At least one of the local farms was known to have brought in wild adult Penaeus indicus from the Red Sea for broodstock, so this could be the origin of the new WSSV variant.
The new TSV genotype found in Saudi Arabia was shown to be distinct from TSV isolates currently found in Southeast Asia and Latin America. This indicated that the source of TSV in Saudi Arabia was probably not an introduction through importation of shrimp, as seen in other areas.
The affected farms are in close proximity to each other, so if a viral disease outbreak occurred on one farm, the disease could easily have spread to nearby farms by sea birds. Both new types of WSSV and TSV were found in the same farms, and they were likely from the same origin, the Red Sea.
Thus, to exclude these viruses, it is recommended to frequently monitor wild broodstock using sensitive diagnostic tools such as PCR to develop specific pathogen-free shrimp and to eventually select for TSV-resistant P. indicus.
(Editor’s Note: This article was originally published in the May/June 2012 print edition of the Global Aquaculture Advocate.)
Authors
-

Kathy F.J. Tang
Department of Veterinary Science and Microbiology
University of Arizona
Tucson, Arizona 85721 USA[117,100,101,46,97,110,111,122,105,114,97,46,117,64,117,121,106,103,110,101,102]
-
Carlos R. Pantoja
Department of Veterinary Science and Microbiology
University of Arizona
Tucson, Arizona 85721 USA -

Donald V. Lightner, Ph.D.
Department of Veterinary Science and Microbiology
University of Arizona
Tucson, Arizona 85721 USA
Tagged With
Related Posts

Health & Welfare
Saudi Arabia developing effective farmed shrimp biosecurity strategy
Biosecurity strategies adopted by the shrimp industry in the Kingdom of Saudi Arabia have resulted in historical production in 2015 and a similar forecast for 2016, and are a significant step towards the long-term goal of sustainable aquaculture.

Health & Welfare
Shrimp biofloc production trials in Saudi Arabia
An R&D trial was conducted with shrimp biofloc technology at a commercial facility in Saudi Arabia. Results indicated that biofloc technology can be effectively applied, with some protocol adjustments, in the KSA’s extreme desert environmental conditions.

Health & Welfare
CENIACUA develops WSSV-resistant shrimp in Colombia
To combat white spot syndrome virus (WSSV) in white shrimp, Corporación Centro de Investigación de la Acuacultura de Colombia (CENIACUA) initiated a selective-breeding program to develop resistance in shrimp.
Health & Welfare
Study: TSV exposure may lessen YHV effects in white shrimp
Since Taura syndrome virus was in the Americas when yellow head virus become pandemic in Asia, it is possible TSV prevented shrimp in the Western Hemisphere from infection by materials from Asia containing YHV. To test this idea, the authors evaluated the viral interactions in shrimp preinfected with TSV and then challenged with YHV.


