Findings indicate that osmoregulation in Baltic cod is a complex process and that western and eastern Baltic cod subpopulations respond differently to salinity changes

The Atlantic cod (Gadus morhua) is a key euryhaline (able to adapt to a wide range of salinities) fish species in the North Atlantic Ocean, with significant commercial and ecological value that will be threatened by the expected climate-related decline in salinity in the Baltic Sea.
Cod adapt to salinity levels ranging from 35 ppt in the North Atlantic and 20 ppt in the surface waters of Kattegat and the western Baltic to about 3 ppt in the northeastern Baltic; thus, they provide an excellent model to study the molecular mechanisms underlying salinity adaptations.
Cod populations in the Baltic Sea have been traditionally divided into two subpopulations (western, WBC; and eastern, EBC) existing in higher- and lower-salinity waters, respectively. In recent decades, both Baltic cod subpopulations have declined massively. One of the reasons for the poor condition of cod in the Baltic Sea is environmental factors, including salinity.
Our preliminary studies showed that Eastern and Western Baltic subpopulations from the natural environment differ in gene expression, which might be related to salinity tolerance. However, understanding of the local genetic adaptation to low salinity in the Baltic Sea is still limited. Further, in recent decades, the Baltic cod population has decreased, especially the EBC, which has experienced a strong depletion manifested by decreased resistance to pathogens, length-weight factor, reproduction/productivity and an increase in natural mortality. The reason for this, alongside overfishing, is the change in climate and environmental conditions, including salinity, which has caused a reduction in spawning areas.
This article – summarized from the original publication (Malachowicz, M. et al. 2023. Diverse Transcriptome Responses to Salinity Change in Atlantic Cod Subpopulations. Cells 2023, 12(23), 2760) – reports on a study that investigated inter-population differences in Atlantic cod responses to salinity changes.
Study setup
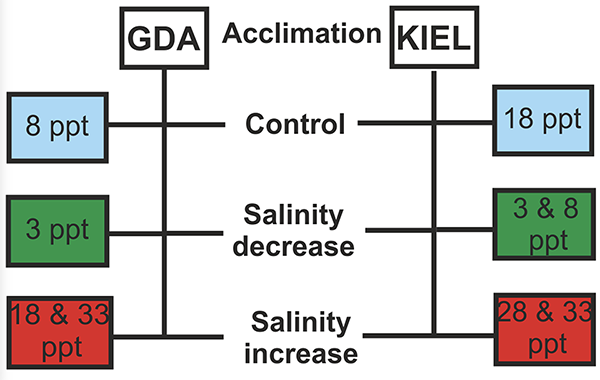
This study was conducted at the marine station of the University of Gdańsk in Hel, Poland. Wild cod juveniles were collected from the Western and Eastern Baltic cod subpopulations and stocked into tanks with recirculated water at the station. Tank water salinity was gradually changed, and then gene expression in gill tissue samples was measured using a molecular technique called genome-wide oligonucleotide DNA microarray (a revolutionary tool for large-scale parallel analyses of genome sequence and gene expression) to characterize the presumed salinity-regulated genes and the biological and molecular processes involved in salinity adaptation.
For detailed information on the experimental design and animal husbandry; RNA extraction and microarray analysis; and other study analyses, refer to the original publication.
Results and discussion
This study compared transcriptomic responses (gene expressions into messenger ribonucleic acid, mRNA, containing the genetic sequence of genes and read in an organism cell in the process of synthesizing specific proteins) to salinity changes in the western and eastern Baltic cod subpopulations represented by fish from the Kiel Bight (KIEL) and the Gulf of Gdańsk (GDA), respectively. The results are in accordance with our previous studies using other molecular techniques.
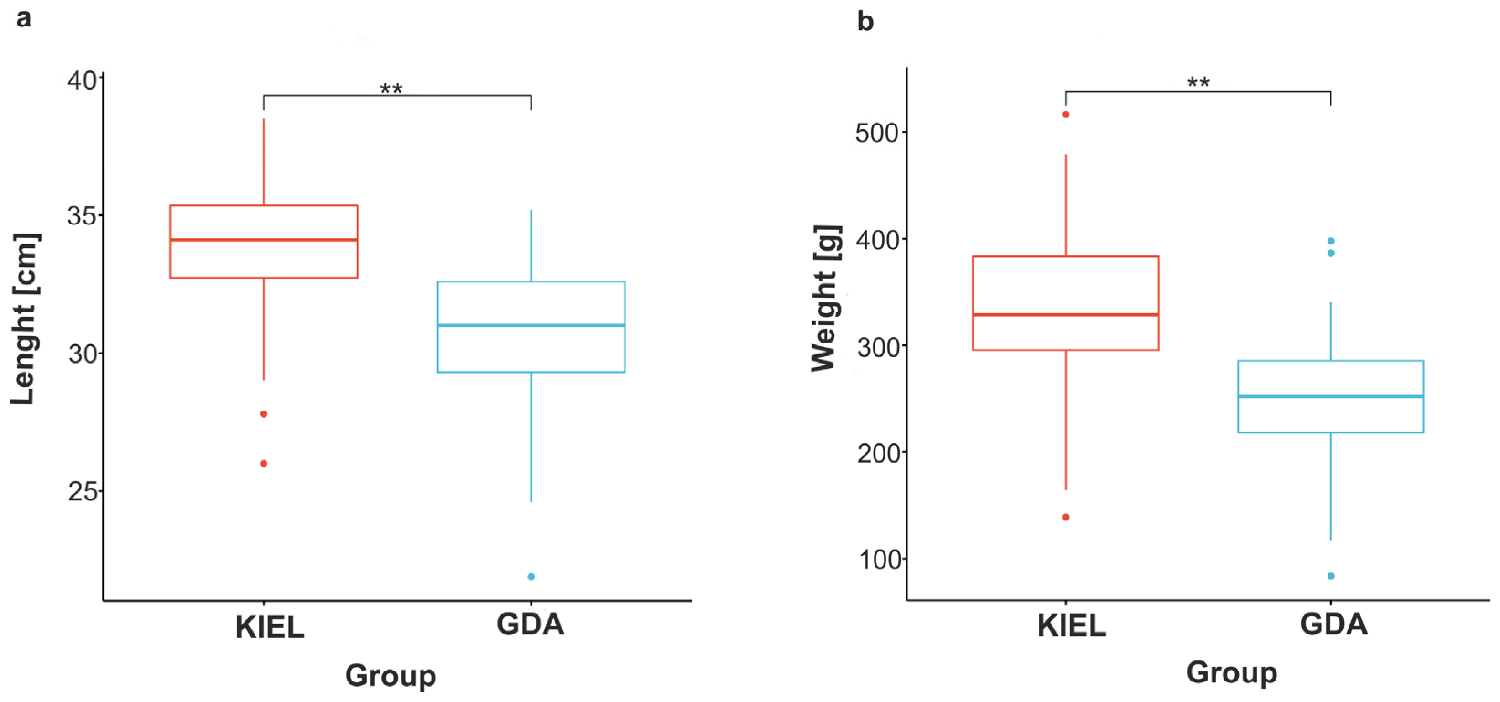
All individuals used in this study were measured in length and weight and showed statistically significant differences between the WBC and EBC subpopulations (Fig. 2). These results showed the poor overall condition of the EBC population, which may cause difficulties in recovery of the stock. A comparison of the magnitude of expression changes between the GDA and KIEL groups revealed that during salinity reductions, the GDA groups responded more robustly; this may indicate that the EBC subpopulation is better positioned for a low salinity future.
Further, we found that these two subpopulations displayed different expression patterns (presumably adaptive responses) to salinity fluctuations. Despite several similarities, 400 differentially expressed genes (DEGs; used to understand the biological differences between healthy and diseased conditions in organisms) showed statistically significant differences between the KIEL and GDA cod groups evaluated at different time points and salinities. These results provide new insights into the molecular mechanisms underlying salinity adaptation in Baltic cod subpopulations.
The analysis of gene expression revealed many subpopulation-dependent genes involved in immune defense in WBC and EBC, thus indicating that salinity fluctuations have a complex effect on the immune system, which is in accordance with previous studies in other bony fishes. Changes in immune-related pathways, in turn, can affect susceptibility to bacterial, viral, and parasitic infections.
Among the subpopulation-dependent genes, there were several gene transcripts belonging to pattern recognition receptors (PRRs), key sensors of the innate (non-specific) immune system of the fish. These results suggest that different types of gene transcripts are activated during salinity fluctuations in Baltic cod. Together, these results suggest that salinity fluctuations affect immune-related genes depending on the Baltic cod subpopulation, which affect their immune responses and, thus, their ability to detect pathogens.
Also, genes involved in cell structure, motility, intercellular communication, and morphogenesis showed differences between the EBC and WBC subpopulations. Additionally, genes involved in keratinization were differentially expressed in the KIEL and GDA groups in response to increased salinity. During the keratinization process, keratins (the key structural material making up scales, hair, nails, feathers, horns, claws, hooves and the outer layer of skin among vertebrates) accumulate inside epithelial tissue cells to form a barrier, thereby reducing water loss during dehydration.
Experts: Climate change is altering fishing industry in North and Baltic seas
Endocrine regulation, including thyroid hormones, is important in salinity acclimatization in euryhaline fish. In this study, thyroid hormone signaling was one of the most enriched pathways, and various gene transcripts were differentially expressed between the EBC and WBC subpopulations. These results suggest that the impacts of salinity on thyroid hormone homeostasis (steady condition) and the activity of the presented genes depend on the cod subpopulation. Our results also revealed that salinity fluctuations cause subpopulation-dependent effects on lipid metabolism in Baltic cod. Lipids provide energy to maintain osmotic balance and regulate membrane structure.
Salinity is an important abiotic factor that affects cod activity, distribution, and reproduction in the Baltic Sea. Due to climate change and increased precipitation in northern latitudes, salinity in the Baltic Sea is expected to decrease. Areas with sea surface salinity less than 7 ppt have expanded since the 1970s and could cover the whole Baltic Sea in the coming decades. Previous studies on the adaptation of Atlantic cod to low salinity in the Baltic Sea showed that the mechanisms may be different in divergent subpopulations of cod, which may contribute to a strong and effective reproductive barrier. Overall, our study results indicate differential metabolic reprogramming in subpopulations of EBC and WBC in response to salinity.
Perspectives
Our study of gene expression analysis showed the complexity of salinity adaptation in Atlantic cod from the Baltic Sea and revealed that Eastern and Western cod subpopulations respond differently to fluctuations in salinity. Among the subpopulation-dependent genes were those involved in the immune system, which could thus affect susceptibility to pathogens.
We also showed that the EBC and WBC subpopulations differ in metabolism reprogramming, gill remodeling, and programmed cell death. This study provides the first general insight into the pathways and functional categories involved in the response of two Baltic cod subpopulations to salinity fluctuations.
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Author
-
Dr. Roman Wenne
Corresponding author
Institute of Oceanology Polish Academy of Sciences, Powstanców Warszawy 55, 81-712 Sopot, Poland[108,112,46,110,97,112,111,105,64,101,110,110,101,119,114]
Related Posts

Intelligence
NASF continues with deep dives into fisheries, aquafeeds and international conflicts
Day two of the North Atlantic Seafood Forum delves into geopolitical conflicts that bleed into fishery management and sustainability initiatives.

Intelligence
As sturgeon farming grows, demand concerns emerge
Caviar, or lightly salted sturgeon roe, has been enjoyed for centuries as an expensive gourmet delicacy. After a drastic decline in wild sturgeon stocks, aquaculture stepped in to fill the void. But can farmed supply find lasting balance with market demand?

Health & Welfare
Atlantic cod genomics and broodstock development project
The Atlantic Cod Genomics and Broodstock Development Project has expanded the gene-related resources for the species in Canada.

Health & Welfare
Photoperiod regulation inhibits spawning, promotes growth in Atlantic cod
Research has shown that an extended photoperiod for Atlantic cod females leads to reduced gonadal growth, faster body growth and shorter grow-out production periods.



