Tests confirmed that a line of P. stylirostris showed no signs of infection 32 days after ingesting IHHNV-infected shrimp tissue

Much research effort has been directed toward the detection and quantification of viral infections in cultured shrimp. Molecular techniques have proven very effective, and determination of the number of viral genomes in infected individuals has become one of the most important means of monitoring the progression of shrimp viral diseases.
Determination of viral loads in other organisms is based on methods that require cell cultures, but reliable methods for maintaining continuous shrimp cell cultures have not yet been achieved. Work at the University of Arizona in Tucson, Arizona, USA, however, has developed a real-time polymerase chain reaction (PCR) method that allows the quantification of viral genomes in individual shrimp without the need for cell cultures.
Real-time PCR
The PCR method involves the addition of a TaqMan probe in a 5’ nuclease assay. In this application, the TaqMan probe is labeled with a reporter dye at the 5’ end and a quencher dye at the 3’ end. The quencher suppresses the fluorescence emitted by the reporter through Forster resonance energy transfer.
During PCR, the Taq polymerase extends the new DNA from its upstream primer. Its 5’ to 3’ exonuclease activity cleaves the reporter dye from the TaqMan probe, which hybridizes to an internal portion of the template DNA. This results in the destruction of the quenching effect and, subsequently, an increase in fluorescence. The Taq polymerase synthesizes amplicons that hybridize to the TaqMan probe such that the amount of fluorescence is in proportion to the number of amplicons generated.
A major feature of real-time PCR is the measurement of DNA amplification at each cycle, as contrasted to traditional PCR, which measures the accumulated amplification at the completion of cycling. In addition, real-time PCR measures fluorescence intensity. This eliminates post-PCR processing, greatly increases throughput, and reduces the risk of contamination.
IHHNV PCR
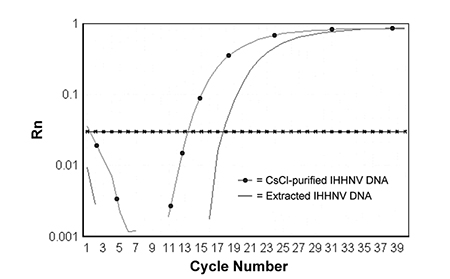
During the development of a PCR assay for infectious hypodermal and hematopoietic necrosis virus, a pair of PCR primers and a TaqMan probe were selected from open reading frame 1 of the IHHNV genome to amplify an 81-bp DNA fragment. The upstream (IHHNV1608F) and downstream (IHHNV1688R) primer sequences are: 5’-TAC TCC GGA CAC CCA ACC A-3’ and 5’-GGC TCT GGC AGC AAA GGT AA-3, respectively. The sequence of TaqMan probe is: 5’-ACC AGA CAT AGA GCT ACA ATC CTC GCC TAT TTG-3’.
Both primers and TaqMan probe were shown to be specific for IHHNV, and can be used to amplify IHHNV from either purified virions or extracted DNA from IHHNV-infected shrimp (Fig. 1).
Quantitative studies
To determine the copy number of the IHHNV genome, a plasmid containing the target IHHNV sequence (designated pIHHNV-P4) was constructed and used as a quantitative standard. A plasmid is a circular, double-stranded unit of DNA that replicates within a cell independently of the chromosomal DNA. Plasmids are most often found in bacteria, and used in recombinant DNA research to transfer genes between cells.
The concentration of pIHHNV-P4 was determined through spectrophotometric analysis, with varying amounts of plasmid used in the IHHNV PCR assay. This method has a detection limit of five copies and linear range up to a billion copies.
Proven results
The assay was used to quantify IHHNV in infected shrimp collected from Mexico, Guam, the Philippines, and Belize, and from laboratory challenge studies. The quantitative analysis showed that wild-caught, large juveniles of Penaeus stylirostris collected in 1996 from the Gulf of California in Mexico were naturally infected with IHHNV and contained up to 10 billion copies of IHHNV per μg of DNA.
Similar quantities of IHHNV were detected in hatchery-raised, small juveniles of P. stylirostris collected from Guam in 1995, and in farm-raised postlarval (P. monodon) from the Philippines in 1996. Laboratory-infected P. stylirostris contained approximately one billion copies of IHHNV 31 days after being fed IHHNV-infected shrimp tissue.
Tests also confirmed that samples of Super Shrimp®, a line of P. stylirostris selected for IHHNV resistance, showed no signs of infection 32 days after ingesting IHHNV-infected shrimp tissue. In P. vannamei with Runt-Deformity Syndrome (RDS) collected in Belize this year, 10 billion copies of IHHNV were detected (Table 1).
Tang, Quantitative analysis of IHHNV, Table 1
| Source | Species | Origin | Tissue | Copy No. μg-1 DNA |
|---|---|---|---|---|
| Mexico | P. stylirostris | Wild-Caught | Gills | 1.1 x 109 |
| Pleopods | 4.2 x 108 | |||
| Guam | P. stylirostris | Hatchery | Heads | 1.2 x 109 |
| Philippines | P. monodon | Farm | Whole Postlarvae | 1.6 x 109 |
| Mexico | P. stylirostris | Lab Infection Day 31, Post-Feeding | Pleopods | 2.2 x 108 |
| Panama | P. stylirostris (Super Shrimp®) | Lab Infection Day 32, Post-Feeding | Heads | Not Detected |
| Belize | P. vannamei | Farm | Pleopods | 4.5 x 109 |
Belize Aquaculture, Ltd. has used real-time PCR to evaluate IHHNV in P. vannamei showing RDS. Ten shrimp each of three sizes (20, 8 and 4 grams) were selected from a pond affected with RDS. Pooled pleo-pods of each group were submitted for real-time PCR analysis.
Results showed the number of copies of IHHNV varied by shrimp size. The smallest shrimp most affected by RDS had the heaviest viral load (1 x 109 copies of IHHNV), the intermediate size had an intermediate load (6.3 x 108), and the largest shrimp had the lightest viral load (4.4 x 107).
Based on these results, real-time PCR analysis was used to help select shrimp from a broodstock pond stocked with large non-runted shrimp from an IHHNV-infected production pond. The majority of shrimp from that pond had 107 to 109 copies of IHHNV, but 10 percent of the broodstock tested had only 103 to 104 copies of IHHNV. These broodstock are now being bred to determine if offspring will also have lower levels of IHHNV, and therefore less tendency to develop runt deformity.
Conclusion
Real-time PCR is a practical technique that can rapidly quantify a large number of samples with high specificity and sensitivity. The use of this method to detect and quantify IHHNV in penaeid shrimp has already revealed new information regarding levels of IHHNV infection and the progress of viral infections.
(Editor’s Note: This article was originally published in the October 2001 print edition of the Global Aquaculture Advocate.)
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Authors
-

Kathy F.J. Tang
Department of Veterinary Science and Microbiology
University of Arizona
Tucson, Arizona, USA -

Donald V. Lightner, Ph.D.
Department of Veterinary Science and Microbiology
University of Arizona
Tucson, Arizona, USA -

Robins P. McIntosh
Belize Aquaculture, Ltd.
Belize City, Belize
Tagged With
Related Posts

Health & Welfare
A study of Zoea-2 Syndrome in hatcheries in India, part 1
Indian shrimp hatcheries have experienced larval mortality in the zoea-2 stage, with molt deterioration and resulting in heavy mortality. Authors investigated the problem holistically.

Health & Welfare
A study of Zoea-2 Syndrome in hatcheries in India, part 2
Indian shrimp hatcheries have experienced larval mortality in the zoea-2 stage, with molt deterioration and resulting in heavy mortality. Authors considered biotic and abiotic factors. Part 2 describes results of their study.

Aquafeeds
Biosecurity protocols needed for shrimp feeds, feeding practices
Shrimp aquafeeds – live, fresh or formulated – should not be an entry point of potential pathogens to the shrimp and/or to their culture systems.

Health & Welfare
Blue alternative: High Health introduces SPF blue shrimp to Thailand
Blue shrimp are very similar to Pacific white shrimp, and can be raised under similar conditions. Blues grow faster and tolerate lower water temperatures.

